Abstract
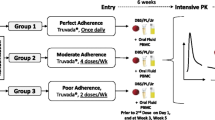
Pre-exposure prophylaxis (PrEP) containing antiretrovirals tenofovir disoproxil fumarate (TDF) or tenofovir alafenamide (TAF) can reduce the risk of acquiring HIV. Concentrations of intracellular tenofovir-diphosphate (TFV-DP) measured in dried blood spots (DBS) have been used to quantify PrEP adherence; although even under directly observed dosing, unexplained between-subject variation remains. Here, we wish to identify patient-specific factors associated with TFV-DP levels. Data from the iPrEX Open Label Extension (OLE) study were used to compare multiple covariate selection methods for determining demographic and clinical covariates most important for drug concentration estimation. To allow for the possibility of non-linear relationships between drug concentration and explanatory variables, the component selection and smoothing operator (COSSO) was implemented. We compared COSSO to LASSO, a commonly used machine learning approach, and traditional forward and backward selection. Training (N = 387) and test (N = 166) datasets were utilized to compare prediction accuracy across methods. LASSO and COSSO had the best predictive ability for the test data. Both predicted increased drug concentration with increases in age and self-reported adherence, the latter with a steeper trajectory among Asians. TFV-DP reductions were associated with increasing eGFR, hemoglobin and transgender status. COSSO also predicted lower TFV-DP with increasing weight and South American countries. COSSO identified non-linear relationships between log(TFV-DP) and adherence, weight and eGFR, with differing trajectories for some races. COSSO identified non-linear log(TFV-DP) trajectories with a subset of covariates, which may better explain variation and enhance prediction. Future research is needed to examine differences identified in trajectories by race and country.


Similar content being viewed by others
Abbreviations
- AIC:
-
Akaike information criterion
- ANOVA:
-
Analysis of variance
- ART:
-
Antiviral therapy
- BIC:
-
Bayesian information criterion
- BLQ:
-
Below levels of quantification
- COSSO:
-
Component selection and smoothing operator
- DBS:
-
Dried blood spots
- eGFR:
-
Estimated glomerular filtration rate
- GAM:
-
Generalized additive model
- HIV:
-
Human immunodeficiency virus
- iPrEx:
-
Spanish: iniciativa profilaxis pre-exposición and English: pre-exposure prophylaxis initiative
- LASSO:
-
Least absolute shrinkage and selection operator
- MSE:
-
Mean square error
- MSM:
-
Men who have sex with men
- OLE:
-
Open label extension
- PBMC:
-
Peripheral blood mononuclear cells
- PrEP:
-
Pre-exposure prophylaxis
- PWH:
-
Persons with HIV
- RMSE:
-
Root mean square error
- SD:
-
Standard deviation
- SS-ANOVA:
-
Smoothing spline analysis of variance
- TAF:
-
Tenofovir alafenamide
- TDF:
-
Tenofovir disoproxil fumarate
- TFV:
-
Tenofovir
- TFV-DP:
-
Tenofovir-diphosphate
References
WHO (2020) HIV/AIDS. 2020. Available from https://www.who.int/news-room/fact-sheets/detail/hiv-aids.
Fauci AS et al (2019) Ending the HIV epidemic: a plan for the United States. JAMA 321(9):844–845
Bacchetti P, Moss AR (1989) Incubation period of AIDS in San Francisco. Nature 338(6212):251–253
Rosenberg PS, Goedert JJ, Biggar RJ (1994) Effect of age at seroconversion on the natural AIDS incubation distribution. Multicenter Hemophilia Cohort Study and the International Registry of Seroconverters. AIDS 8(6):803–10
Hosek SG et al (2013) The acceptability and feasibility of an HIV preexposure prophylaxis (PrEP) trial with young men who have sex with men. J Acquir Immune Defic Syndr 62(4):447–456
Van Damme L et al (2012) Preexposure prophylaxis for HIV infection among African women. N Engl J Med 367(5):411–422
Fonner VA et al (2016) Effectiveness and safety of oral HIV preexposure prophylaxis for all populations. AIDS 30(12):1973–1983
Grant RM et al (2014) Uptake of pre-exposure prophylaxis, sexual practices, and HIV incidence in men and transgender women who have sex with men: a cohort study. Lancet Infect Dis 14(9):820–829
Marrazzo JM et al (2015) Tenofovir-based preexposure prophylaxis for HIV infection among African women. N Engl J Med 372(6):509–518
Van der Straten A et al (2012) Unraveling the divergent results of pre-exposure prophylaxis trials for HIV prevention. AIDS 26(7):F13–F19
Baeten JM et al (2012) Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. N Engl J Med 367(5):399–410
Thigpen MC et al (2012) Antiretroviral preexposure prophylaxis for heterosexual HIV transmission in Botswana. N Engl J Med 367(5):423–434
Anderson PL et al (2012) Emtricitabine-tenofovir concentrations and pre-exposure prophylaxis efficacy in men who have sex with men. Sci Transl Med 4(151):151125
Koss CA et al (2017) Differences in cumulative exposure and adherence to tenofovir in the VOICE, iPrEx OLE, and PrEP demo studies as determined via hair concentrations. AIDS Res Hum Retrovir 33(8):778–783
CDC (2017) HIV and Gay and Bisexual Men. Available from https://www.cdc.gov/hiv/group/msm/index.html.
CDC (2020) PrEP. Available from https://www.cdc.gov/hiv/basics/prep.html.
Brooks KM, Anderson PL (2018) Pharmacologic-based methods of adherence assessment in HIV prevention. Clin Pharmacol Ther 104(6):1056–1059
Anderson PL et al (2018) Intracellular tenofovir-diphosphate and emtricitabine-triphosphate in dried blood spots following directly observed therapy. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.01710-17
Yager J et al (2020) Intracellular tenofovir-diphosphate and emtricitabine-triphosphate in dried blood spots following tenofovir alafenamide: the TAF-DBS Study. J Acquir Immune Defic Syndr 84(3):323–330
Sethuraman VS et al (2007) Sample size calculation for the Power Model for dose proportionality studies. Pharm Stat 6(1):35–41
Smith BP et al (2000) Confidence interval criteria for assessment of dose proportionality. Pharm Res 17(10):1278–1283
Coyle RP et al (2020) Factors associated with tenofovir diphosphate concentrations in dried blood spots in persons living with HIV. J Antimicrob Chemother 75(6):1591–1598
Olanrewaju AO et al (2020) Enzymatic assay for rapid measurement of antiretroviral drug levels. ACS Sens 5(4):952–959
Grant RM et al (2010) Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med 363(27):2587–2599
Breiman L (1995) Better subset regression using the nonnegative garrote. Technometrics 37(4):373–384
Bzdok D, Altman N, Krzywinski M (2018) Statistics versus machine learning. Nat Methods 15(4):233–234
Tibshirani R (1996) Regression shrinkage and selection via the lasso. J R Stat Soc 58(1):267–288
Lin Y, Zhang HH (2006) Component selection and smoothing in multivariate nonparametric regression. Ann Stat 34(5):2272–2297
Grohskopf LA et al (2013) Randomized trial of clinical safety of daily oral tenofovir disoproxil fumarate among HIV-uninfected men who have sex with men in the United States. J Acquir Immune Defic Syndr 64(1):79–86
Kumar VS, Webster M (2016) Measuring adherence to HIV pre-exposure prophylaxis through dried blood spots. Clin Chem 62(7):1041–1043
Vittinghoff E et al (2014) Regression methods in biostatistics, linear, logistic, survival and repeated measures models, 2nd edn. Springer, New York
Harrell F (2015) Regression modeling strategies, 2nd edn. Springer, Cham
Heinze G, Wallisch C, Dunkler D (2018) Variable selection—a review and recommendations for the practicing statistician. Biom J 60(3):431–449
Lockhart R et al (2014) A significance test for the lasso. Ann Stat 42(2):413–468
Tibshirani RJ et al (2016) Exact post-selection inference for sequential regression procedures. J Am Stat Assoc 111(514):600–620
Tibshirani R et al (2019) selectiveinference: tools for post-selection inference. Available from https://CRAN.R-project.org/package=selectiveInference.
Wang Y et al (1997) Using smoothing spline anova to examine the relation of risk factors to the incidence and progression of diabetic retinopathy. Stat Med 16(12):1357–1376
Zhang HH, Lin C (2013) cosso: fit regularized nonparametric regression models using COSSO penalty. R package version 2.1–1. Available from https://CRAN.R-project.org/package=cosso.
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33(1):1–22
Venables WN, Ripley BD (2002) Modern applied statistics with S, 4th edn. Springer, New York
Wood SN (2017) Generalized additive models: an introduction with R, 2nd edn. CRC Press, Boca Raton
Efron E et al (2004) Least angle regression. Annals of Statistics 32(2):407–499
Grant RM et al (2020) Sex hormone therapy and tenofovir diphosphate concentration in dried blood spots: primary results of the iBrEATHe study. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa1160
Zheng JH et al (2016) Application of an intracellular assay for determination of tenofovir-diphosphate and emtricitabine-triphosphate from erythrocytes using dried blood spots. J Pharm Biomed Anal 122:16–20
Huang J, Horowitz JL, Wei F (2010) Variable selection in nonparametric additive models. Ann Stat 38(4):2282–2313
Storlie CB et al (2011) Surface estimation, variable selection, and the nonparametric oracle property. Stat Sin 21(2):679–705
Bertsimas D, King A, Mazumder R (2016) Best subset selection via a modern optimization lens. Ann Stat 44(2):813–852
Hastie T, Tibshirani R, Tibshirani R (2020) Best subset, forward stepwise or lasso? Analysis and recommendations based on extensive comparisons. Stat Sci 35(4):579–592
Daume H (2004) From zero to reproducing Kernel Hilbert spaces in twelve pages or less. Available from http://users.umiacs.umd.edu/~hal/docs/daume04rkhs.pdf.
Acknowledgements
This works was funded by NIH Grants UO1 AI64002, UO1 AI84735 and RO1 AI122298. The authors would like to acknowledge the iPrEX OLE study team and volunteers who participated in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Camille M. Moore and Samantha MaWhinney are the joint senior.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
Statistical methods
Simulation studies have shown the relative performance of stepwise selection procedures and LASSO depend on the levels of correlation between the independent variables considered, the number of non-zero regression coefficients, sample size, and the signal to noise ratio [42, 45, 47, 48]. Here we summarize the methods considered to assess our goal to compare and contrast these methods in a real-world application and to understand if COSSO, which allows for non-linear relationships, offered additional insights into factors associated with TFV-DP levels.
Backward/forward
In least squares linear regression, the residual sum of squares (RSS) is minimized to produce the best fit to the data. Let \(i = \{1,\dots ,n\}\) be the number of subjects and \(j = \{1,\dots ,k\}\) be the number of covariates. \({y}_{i}\) is the outcome and \({{\varvec{x}}}_{i}\) is the covariate vector for subject i. \({\beta }_{j}\) is the coefficient for each covariate and \({\beta }_{0}\) is the intercept. The RSS is:
Forward and backward model selection methods use the RSS solutions, evaluating each covariate one at a time using p-values or model fit statistics such as R-squared, Akaike information criterion (AIC) or Bayesian information criterion (BIC). We used AIC for the forward and backward selection methods. Lower AIC values indicate better model fit.
The forward selection method starts with no covariates in the model and AIC is calculated for this initial, intercept-only model. Then, a set of models is fit by adding each covariate individually to the initial model; AIC is calculated for each model and compared to the initial model’s AIC. The covariate that results in the largest reduction in AIC is added to the model. This process is repeated until the inclusion of additional covariates no longer results in a reduction in AIC. Backward selection starts with all potential covariates included in the initial model. A set of models is fit removing each covariate individually from the initial model; again, AIC is calculated and compared to the initial model. The covariate that results in the largest reduction in AIC is removed from the model. This process is repeated until the removal of covariates no longer results in AIC reductions.
Performing model selection based on AIC is related to model selection criteria based on p-value thresholds. It has been shown that when comparing two hierarchically nested models that differ by 1 degree of freedom (i.e. a single variable) in stepwise model selection, using AIC optimization is equivalent to performing model selection using a p-value threshold of 0.157. See Heinze et al. for full details [33]. Two problems arise from these step-wise model selection methods: (1) multiple statistical tests are performed without correction for multiple comparisons, which may lead to a large number of covariates being included in the model, potential overfitting, less generalizability to other datasets, and (2) the relevance of each individual covariate is not tested; rather we test each individual covariate given the set of covariates that has already been selected, so the inclusion or exclusion of a covariate is highly dependent on previous covariate selections in the step-wise process. This may cause small changes to the dataset, such as the exclusion of a single observation, to result in a different set of covariates being chosen. Another major disadvantage to these methods is it only allows for linear relationships.
LASSO
LASSO is a common method used for covariate selection. LASSO also relies on the RSS but adds an additional constraint on \({\varvec{\beta}}\) as follows.
Or equivalently
where \(\lambda \ge 0\) and \(t \ge 0\) are tuning parameters. If \(t_{0} = \sum \left| {\beta_{j}^{0} } \right|\), where \(\beta_{j}^{0}\) is the full least squares estimates, then any values of \(t < t_{0}\) will cause shrinkage of the solutions towards 0, and some coefficients will be exactly 0. Those that are shrunk to 0 are the covariates not deemed important in the model [27]. Going from Eqs. (3) to (4) uses the Lagrange multiplier, and \(\lambda\) and \(t\) have a relation that is dependent on the data.
LASSO is more stable than backward and forward selection so small changes in the data result in similar models selected. The disadvantage of LASSO, especially in the context of our example data, is that it only allows for linear relationships. In addition, LASSO estimates are shrunk towards zero, which leads to increased bias in exchange for better predicted fits [34].
COSSO
COSSO is an extension of smoothing spline ANOVA (SSANOVA), which allows for both non-linear relationships and covariate selection. Standard implementations do not provide significance testing. COSSO utilizes elements of several statistical methods including penalized splines, nonnegative garrote, and SSANOVA.
SSANOVA
The Smoothing Spline ANOVA is a model using a penalized likelihood to allow for non-linear relationships and smoothing of noisy covariate and outcome data. Consider the model
where \({\varvec{x}}_{i} = \left( {{\varvec{x}}_{i}^{\left( 1 \right)} , \ldots , {\varvec{x}}_{i}^{\left( k \right)} } \right)\) is a k-dimensional vector of covariates for subject i, \({f}_{j}\) is the main effect of covariate j and \({f}_{jq}\) is the interaction of covariates j and q where \(q = \left\{ {1, \ldots ,k} \right\}\) the same as\(j\). \({f}_{j}()\) is a smooth function estimate modeled with penalized splines, giving SS-ANOVA flexibility by allowing for non-linear relationships [37]. SSANOVA utilizes a penalty log-likelihood, denoted by\(L(y,f)\), similar to LASSO:
where \({\lambda }_{j}\) is the tuning parameter for each covariate and \(J_{j} \left( {f_{j} } \right) = \mathop \smallint \limits_{0}^{1} \left( {f_{j}^{\prime \prime} \left( {x_{j} } \right)} \right)^{2} dx_{j}\) is the penalty functional. As \({\lambda }_{j}\) tends to infinity \({f}_{j}\) tends to a linear function.
COSSO
Lin and Zhang proposed COSSO to give both flexibility through splines and covariate selection [28]. COSSO is an extension of SSANOVA adding a different penalty term, \(J\left(f\right)\), using a penalized least squares method. The penalty is a sum of component norms rather than the squared component norms used in SS-ANOVA. COSSO reduces to a LASSO when COSSO is used in linear models.
COSSO minimizes the following equation to estimate model parameters:
where, \({\tau }_{n}\) is a smoothing parameter. The penalty term \(J\left(f\right)\) is the sum of Reproducing Kernel Hilbert Space norms (RKHS); the norm in the RKHS \(F\) is denoted by ||.||. Specifically, \({P}^{\alpha }f\) is the orthogonal projection of \(f\) onto \({F}^{\alpha }\) where \(\alpha = \left\{1,\dots ,p\right\}\) and \(p\) is the number of covariates. RKHS are commonly used to control smoothness of functions used in regression [49].
An equivalent form of Eq. (6) that is computationally more efficient is minimizing
where \({\lambda }_{0}\) is a fixed constant and \(\lambda\) is a smoothing parameter. This equation is solved for \(f \in F\) and \({\varvec{\theta}} = \left( {\theta_{1} , \ldots , \theta_{p} } \right)^{T}\) where each \(\theta \ge 0\). From the smoothing spline literature it is well known the solution \(f\) has the form \(f\left( x \right) = \sum\nolimits_{k = 1}^{K} {{\varvec{c}}_{{\varvec{i}}} {\varvec{R}}_{{\varvec{\theta}}} \left( {{\varvec{x}}_{{\varvec{i}}} ,\user2{ x}} \right) + b_{{}} }\), where \({\varvec{c}} = \left\{ {c_{1} , \ldots , c_{K} } \right\}^{T}\) \(\in {\varvec{R}}^{n}\), \(b \in {\varvec{R}}\), and \(R_{\theta } = \sum\nolimits_{\alpha = 1}^{p} {\theta_{\alpha } {\varvec{R}}_{\alpha } }\), with \({\varvec{R}}_{\alpha}\) being the reproducing kernel of \({\varvec{F}}^{\alpha}\). K is the number of spline knots, which cannot exceed the number of subjects in the dataset, \(b\) is the intercept, and \({\varvec{c}}\) is the vector of coefficients for the smooth function\(f\). \({\varvec{\theta}}\) is the shrinkage vector to allow for covariate selection; as \(\lambda\) goes toward 0, the far-right term in the Eq. (7) goes to 0 causing less shrinkage in the \(\theta\) coefficients. COSSO is fit using an iterative procedure. It first solves for the regression parameters (\({\varvec{c}}\) and \(b\)) using the known SSANOVA solution while fixing \({\varvec{\theta}}.\) Then the regression parameters are fixed to solve for the \({\varvec{\theta}}\) using a non-negative garrote, which is a precursor to LASSO [25]. These steps are then iterated until convergence. A single iteration beyond initialization steps has been found to produce estimates with similar accuracy to the fully iterated procedure; therefore, a one-step update procedure is used in practice. To set tuning parameters, \({\lambda }_{0}\) can be fixed at any positive value, and then \(\lambda\) is tuned accordingly using generalized cross-validation or cross validation.
Influence of the number of cross-validation folds and spline knots on COSSO model stability
COSSO relies on cross validation to determine tuning parameters; however, random data partitioning in cross validation may result in the selection of different tuning parameters, and therefore the selection of different covariate subsets, in independent runs of COSSO. In addition, COSSO requires the user to specify the number of knots for the smoothing splines. We investigated the stability of tuning parameter and covariate subset selection to the number of cross validation folds and number of spline knots. While fivefold cross-validation is commonly used for covariate selection techniques such as LASSO or COSSO, our dataset had several categorical variables with a low number of subjects in some categories. Therefore, it was possible for all or most of the subjects in these small categories to be excluded from model fitting in certain data partitions during cross-validation with a low number of folds due to the random sub-sampling. Therefore, we compared cross validation using 20 folds vs. 100 folds in our full dataset (N = 553) and performed a limited exploration of fivefold cross-validation. We were not able to evaluate leave one out cross validation, as the COSSO R package only allows for N/2 folds, where N is the total sample size. We compared results using 45 (minimum allowed by the COSSO R package), 100, and 553 (maximum allowed by the COSSO R package) spline knots. fivefold cross validation was only evaluated using 553 knots for the reasons noted above. We repeated the random sub-sampling 100 times and evaluated model stability in terms of the distribution of the tuning parameter across runs, the number of times the most common covariate subset was chosen, and the total number of different models that were selected across the 100 runs. Model stability generally increased with more spline knots and a larger number of cross validation folds. At 45 knots and 20-folds, the most common model was chosen six times out of 100, while at 553 knots and 100-folds the most common model was chosen 80 times out of 100.
Rights and permissions
About this article
Cite this article
Peterson, S., Ibrahim, M., Anderson, P.L. et al. A comparison of covariate selection techniques applied to pre-exposure prophylaxis (PrEP) drug concentration data in men and transgender women at risk for HIV. J Pharmacokinet Pharmacodyn 48, 655–669 (2021). https://doi.org/10.1007/s10928-021-09763-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-021-09763-y