Abstract
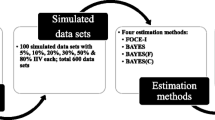
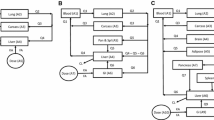
First-order conditional estimation (FOCE) has been the most frequently used estimation method in NONMEM, a leading program for population pharmacokinetic/pharmacodynamic modeling. However, with growing data complexity, the performance of FOCE is challenged by long run time, convergence problem and model instability. In NONMEM 7, expectation–maximization (EM) estimation methods and FOCE with FAST option (FOCE FAST) were introduced. In this study, we compared the performance of FOCE, FOCE FAST, and two EM methods, namely importance sampling (IMP) and stochastic approximation expectation–maximization (SAEM), utilizing the rich pharmacokinetic data of oxfendazole and its two metabolites obtained from the first-in-human single ascending dose study in healthy adults. All methods yielded similar parameter estimates, but great differences were observed in parameter precision and modeling time. For simpler models (i.e., models of oxfendazole and/or oxfendazole sulfone), FOCE and FOCE FAST were more efficient than EM methods with shorter run time and comparable parameter precision. FOCE FAST was about two times faster than FOCE but it was prone to premature termination. For the most complex model (i.e., model of all three analytes, one of which having high level of data below quantification limit), FOCE failed to reliably assess parameter precision, while parameter precision obtained by IMP and SAEM was similar with SAEM being the faster method. IMP was more sensitive to model misspecification; without pre-systemic metabolism, IMP analysis failed to converge. With parallel computing introduced in NONMEM 7.2, modeling speed increased less than proportionally with the increase in the number of CPUs from 1 to 16.



Similar content being viewed by others
References
Sheiner LB, Beal SL (1980) Evaluation of methods for estimating population pharmacokinetics parameters. I. Michaelis-Menten model: routine clinical pharmacokinetic data. J Pharmacokinet Biopharm 8(6):553–571. https://doi.org/10.1007/BF01060053
Beal S, Sheiner L (1998) NONMEM users guide-Part VII: Conditional estimation methods. NONMEM Project Group, San Francisco
Bauer RJ (2017) NONMEM users guide-Introduction to NONMEM 7.4.1. ICON Plc, Dublin
Bauer RJ, Guzy S, Ng C (2007) A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J 9(1):E60-83. https://doi.org/10.1208/aapsj0901007
Gibiansky L, Gibiansky E, Bauer R (2012) Comparison of Nonmem 7.2 estimation methods and parallel processing efficiency on a target-mediated drug disposition model. J Pharmacokinet Pharmacodyn 39(1):17–35. https://doi.org/10.1007/s10928-011-9228-y
Liu X, Wang Y (2016) Comparing the performance of FOCE and different expectation-maximization methods in handling complex population physiologically-based pharmacokinetic models. J Pharmacokinet Pharmacodyn 43(4):359–370. https://doi.org/10.1007/s10928-016-9476-y
Johansson AM, Ueckert S, Plan EL, Hooker AC, Karlsson MO (2014) Evaluation of bias, precision, robustness and runtime for estimation methods in NONMEM 7. J Pharmacokinet Pharmacodyn 41(3):223–238. https://doi.org/10.1007/s10928-014-9359-z
Duffull SB, Kirkpatrick CM, Green B, Holford NH (2005) Analysis of population pharmacokinetic data using NONMEM and WinBUGS. J Biopharm Stat 15(1):53–73. https://doi.org/10.1081/bip-200040824
Plan EL, Maloney A, Mentre F, Karlsson MO, Bertrand J (2012) Performance comparison of various maximum likelihood nonlinear mixed-effects estimation methods for dose-response models. AAPS J 14(3):420–432. https://doi.org/10.1208/s12248-012-9349-2
Chan PL, Jacqmin P, Lavielle M, McFadyen L, Weatherley B (2011) The use of the SAEM algorithm in MONOLIX software for estimation of population pharmacokinetic-pharmacodynamic-viral dynamics parameters of maraviroc in asymptomatic HIV subjects. J Pharmacokinet Pharmacodyn 38(1):41–61. https://doi.org/10.1007/s10928-010-9175-z
An G, Murry DJ, Gajurel K, Bach T, Deye G, Stebounova LV, Codd EE, Horton J, Gonzalez AE, Garcia HH, Ince D, Hodgson-Zingman D, Nomicos EYH, Conrad T, Kennedy J, Jones W, Gilman RH, Winokur P (2019) Pharmacokinetics, Safety, and Tolerability of Oxfendazole in Healthy Volunteers: a Randomized, Placebo-Controlled First-in-Human Single-Dose Escalation Study. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.02255-18
Bauer RJ (2016) Technical guide on various methods in NONMEM 7. NONMEM Plc, Dublin
Almquist J, Leander J, Jirstrand M (2015) Using sensitivity equations for computing gradients of the FOCE and FOCEI approximations to the population likelihood. J Pharmacokinet Pharmacodyn 42(3):191–209. https://doi.org/10.1007/s10928-015-9409-1
Beal SL (2001) Ways to fit a PK model with some data below the quantification limit. J Pharmacokinet Pharmacodyn 28(5):481–504
Boeckmann AJ, Beal SL, Sheiner LB (2017) NONMEM users guide-Part IV: NM-TRAN guide. ICON Plc, Dublin
Bach T, Murry DJ, Stebounova LV, Deye G, Winokur P, An G (2021) Population pharmacokinetic model of oxfendazole and metabolites in healthy adults following single ascending doses. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.02129-20
Bauer RJ (2019) NONMEM tutorial part II: estimation methods and advanced examples. CPT Pharmacometrics Syst Pharmacol. https://doi.org/10.1002/psp4.12422
Acknowledgements
This work was supported by the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health through the Vaccine and Treatment Evaluation Unit (Contracts No. HHSN272200800008C and HHSN24220130020I) and by the National Center for Advancing Translational Sciences grant to the University of Iowa (Grant No. 5U54TR001356) for the work done in the Clinical Research Unit.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bach, T., An, G. Comparing the performance of first-order conditional estimation (FOCE) and different expectation–maximization (EM) methods in NONMEM: real data experience with complex nonlinear parent-metabolite pharmacokinetic model. J Pharmacokinet Pharmacodyn 48, 581–595 (2021). https://doi.org/10.1007/s10928-021-09753-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-021-09753-0