Abstract
We present competitive and uncompetitive drug–drug interaction (DDI) with target mediated drug disposition (TMDD) equations and investigate their pharmacokinetic DDI properties. For application of TMDD models, quasi-equilibrium (QE) or quasi-steady state (QSS) approximations are necessary to reduce the number of parameters. To realize those approximations of DDI TMDD models, we derive an ordinary differential equation (ODE) representation formulated in free concentration and free receptor variables. This ODE formulation can be straightforward implemented in typical PKPD software without solving any non-linear equation system arising from the QE or QSS approximation of the rapid binding assumptions. This manuscript is the second in a series to introduce and investigate DDI TMDD models and to apply the QE or QSS approximation.
Similar content being viewed by others
References
Ariëns EJ, Van Rossum JM, Simonis AM (1957) Affinity, intrinsic activity and drug interactions. Pharmacol Rev 9(2):218–236
Banks HT (1975) Modeling and control in biomedical sciences, lecture notes in biomathematics. Springer, Berlin
Koch G, Schropp J, Jusko WJ (2016) Assessment of non-linear combination effect terms for drug–drug interactions. J Pharmacokinet Pharmacodyn 43(5):461–479
Levy G (1994) Pharmacologic target-mediated drug disposition. Clin Pharmacol Ther 56(3):248–252
Mager DE, Jusko WJ (2001) General pharmacokinetic model for drugs exhibiting target-mediated drug disposition. J Pharmacokinet Pharmacodyn 28(6):507–532
Koch G, Jusko WJ, Schropp J (2017) Target mediated drug disposition with drug–drug interaction, Part I: single drug case in alternative formulations. J Pharmacokinet Pharmacodyn. doi:10.1007/s10928-016-9501-1
Mager DE, Krzyzanski W (2005) Quasi-equilibrium pharmacokinetic model for drugs exhibiting target-mediated drug disposition. Pharm Res 22(10):1589–1596
Gibiansky L, Gibiansky E, Kakkar T, Ma P (2008) Approximations of the target-mediated drug disposition model and identifiability of model parameters. J Pharmacokinet Pharmacodyn 35(5):573–591
Yan X, Chen Y, Krzyzanski W (2012) Methods of solving rapid binding target-mediated drug disposition model for two drugs competing for the same receptor. J Pharmacokinet Pharmacodyn 39(5):543–560
Copland RA (2005) Evaluation of enzyme inhibitors in drug discovery, A guide for medicinal chemists and pharmacologists. Wiley, Hoboken
Peletier LA, Gabrielsson J (2012) Dynamics of target-mediated drug disposition: characteristic profiles and parameter identification. J Pharmacokinet Pharmacodyn 39(5):429–451
Peletier LA, Gabrielsson J (2013) Dynamics of target-mediated drug disposition: how a drug reaches its target. Comput Geosci 17:599–608
Lipton SA (2006) Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat Rev Drug Discov 5(2):160–170
Fenichel N (1979) Geometric singular perturbation theory for ordinary differential equations. J Diff Equ 31:54–98
Vasileva AB (1963) Asymptotic behaviour of solutions to certain problems involving nonlinear differential equations containing a small parameter multiplying the highest derivatives. Russ Math Surv 18:13–83
D’Argenio DZ, Schumitzky A, Wang X (2009) ADAPT 5 user’s guide: pharmacokinetic / pharmacodynamic systems analysis software. Biomedical Simulations Resource, Los Angeles
Beal S, Sheiner LB, Boeckmann A, Bauer RJ (2009) NONMEM user’s guides. Icon Development Solutions, Ellicott City
R Core Team (2014) R: A language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. http://www.R-project.org/
MATLAB Release (2014b) The MathWorks. Inc, MathWorks, Natick
Brenan KE, Campbell SL, Petzold LR (1996) Numerical solution of initial value problems in differential-algebraic equations. Classics in Applied Mathematics, 14 SIAM
Nahorski SR, Ragan CI, Challiss RA (1991) Lithium and the phosphoinositide cycle: an example of uncompetitive inhibition and its pharmacological consequences. Trends Pharmacol Sci 12(8):297–303
Cornish-Bowden A (1986) Why is uncompetitive inhibition so rare? A possible explanation, with implications for the design of drugs and pesticides. FEBS Lett 203(1):3–6
Acknowledgements
This work was supported in part by NIH Grant GM24211.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendices
Appendix 1: Derivation of the final QE and QSS approximation in free concentration variables
Competitive DDI
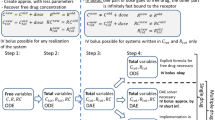
Step 1: Total concentration formulation
Similar to the single drug case [6] the key for the QE or QSS approximation is to reformulate Eqs. (1)–(5) in total drug and total receptor concentration variables. With
we obtain
The baseline initial values are
for \(X \in \{ A,B \}\). The values \(C_A^0, \, C_B^0, \, R^0, \, RC_A^0, \, RC_B^0\) in Eq. (59) are chosen according to Eqs. (6)–(8) and the input functions in Eqs. (54)–(55) according to Eq. (9). Substituting free variables in Eqs. (54)–(58) with total variables from Eqs. (51)–(53) we obtain
In comparison to Eqs. (1)–(5), Eqs. (60)–(64) have the advantage that the parameters \(k_{onX}\) and \(k_{offX}\) appear in the equations of the complexes only.
Step 2: QE and QSS binding relations
We assume rapid binding between \(C_A\) and R, as well as \(C_B\) and R. Hence, QE or QSS approximation of the complexes \(RC_A\) and \(RC_{B}\) in Eqs. (57)–(58) provide the algebraic equations
for \(Y \in \{ D, SS \}\) with Eq. (19) (see Appendices 2, 3). The differential algebraic equation (DAE) form in total variables is then given by Eqs. (60)–(62), (65)–(66).
Step 3: QE and QSS model equations
To avoid solving the coupled non-linear equation system Eqs. (65)–(66) numerically, we transform Eqs. (54)–(56), (65)–(66) back to the free variables. From Eqs. (65)–(66) we obtain the complexes
The next step is to differentiate Eq. (67) and to express \(\frac{d}{dt} C_{totA}, \frac{d}{dt} C_{totB}, \frac{d}{dt} R_{tot}\) appearing at the left hand side of Eqs. (54)–(56) in terms of \(C_A, C_B\) and R and their derivatives. Using Eqs. (51)–(53) we can calculate from Eqs. (54)–(56)
The equivalent matrix form reads
with
Eq. (71) is equivalent to
where
\(Q^{-1}\) denotes the inverse matrix of Q and the explicit representation of \(M_{Com}\) is listed in Table 1.
Uncompetitive DDI
Step 1: Total concentration formulation
The total drug and receptor variables are
and we obtain
The baseline initial values are obtained by applying Eqs. (73)–(75) to the initial values Eqs. (28)–(31). This leads to
and the input functions Eqs. (32)–(33).
Again substituting the free variables in Eqs. (76)–(80) yields
Note that in the formulation Eqs. (81)–(85) the parameter \(k_{onX}, \, k_{offX}\), intended for elimination show up in the equations of the complexes only.
Step 2: QE binding relations
In Appendix 2 it is shown that the QE approximation provides the algebraic equations
and the resulting DAE consists of Eqs. (81)–(83), (86), (87).
Step 3: QE model equations
Using Eqs. (73)–(75) and Eqs. (76)–(78) we can compute
In addition, from Eqs. (86)–(87) we obtain by differentiation
With Eqs. (88)–(92) the equivalent matrix form reads
with \(P(C_A,C_B ,R) = I+\hat{P}(C_A,C_B,R)\),
and
Finally, Eq. (93) can be written as explicit ODE
where
is listed in Table 1.
Appendix 2: QE approximation
The QE approximation is based on the theory of Fenichel [14] which allows a specific selection of the rates to be accelerated.
Competitive
To justify the QE approximation we increase the binding rates \(k_{onX},k_{offX}\), where \(X \in \{A,B\}\), by replacing with \(\frac{1}{\varepsilon } k_{onX}\), \(\frac{1}{\varepsilon } k_{offX}\) with \(\varepsilon > 0\) small in Eqs. (54)–(58). Since the new constants are much larger this can be regarded as rapid binding and we obtain
Multiplying Eqs. (94)–(95) by \(\varepsilon\) gives
Taking the limit \(\varepsilon \rightarrow 0\) in Eqs. (96)–(97) results in
Dividing Eq. (98) by \(k_{onA}\) and Eq. (99) by \(k_{onB}\) gives the QE approximation of the complexes
Uncompetitive
Accelerating the binding rates \(k_{onX}\) and \(k_{offX}\) with \(X \in \{A,AB\}\) in Eqs. (76)–(80) gives
Multiplying Eqs. (102)–(103) by \(\varepsilon\) leads to
Taking the limit \(\varepsilon \rightarrow 0\) in Eqs. (104)–(105) results in
Substituting Eq. (107) in Eq. (106) leads to
Dividing Eq. (108) with \(k_{onA}\) and Eq. (109) with \(k_{onAB}\) gives the QE approximation of the complexes
Appendix 3: QSS approximation
Following the classical singular perturbation theory [15] all complex related processes are assumed to be rapid, including the internalization from the complexes.
Competitive
Accelerating the rates with \(\varepsilon\) small in Eqs. (54)–(58) yields
Multiplying Eqs. (112)–(113) by \(\varepsilon\) and taking the limit \(\varepsilon \rightarrow 0\)
Hence, the QSS approximation reads
Uncompetitive
We obtain from Eqs. (76)–(80) with \(\varepsilon\) small
Multiplying these equations by \(\varepsilon\) and then taking the limit \(\varepsilon \rightarrow 0\) results in
Inserting Eq. (122) in Eq. (121) gives
Dividing Eq. (123) by \(k_{onA}\) and Eq. (124) by \(k_{onAB}\) provides
Appendix 4: Baseline initial values for the uncompetitive TMDD model
According to Eqs. (26)–(27) the baseline conditions for the complexes with the concentrations \(C_A^0, C_B^0 \ge 0\) are
Applying Cramer’s rule to Eq. (127) and using the definition from Eq. (19) yields the solution
Inserting Eqs. (128)–(129) into the baseline condition of the receptor equation (78) leads to
which is equivalent to
The baseline concentrations of the input functions then follow from Eqs. (76)–(77).
Appendix 5: Source codes
The matrix representation applied in Eqs. (14)–(15) and Eqs. (43)–(44) is of the general form
Hence, performing matrix multiplication the right hand side of the differential equation reads
compare the lines 113–128 for the competitive and the lines 221–239 for the uncompetitive case. The variables \(H_1\),...,\(H_3\) correspond to DADT(1), ..., DADT(3) in NONMEM and XP(1), ..., XP(3) in ADAPT 5.
The lines of the code are numbered for referencing but are not part of the code implementation.
NONMEM control stream for competitive DDI TMDD
The $DES block of the control stream is presented. Additionally, the first lines of the data file is shown to present the IV infusion mechanism. The full control stream is available in the supplemental material.
101: $DES
102: EPSILON = 1e-4
103: ; Dose at T1 = 0
104: INA = 0
105: INB = 0
106: IF (T.GE.0.AND.T.LE.0+EPSILON) THEN
107: INA = 100*EPSILON**(−1)
108: INB = 100*EPSILON**(-1)
109: ENDIF
110: CA = A(1)/V
111: CB = A(2)/V
112: R = A(3)
113: DET = R**2+CA*KDB+CB*KDA+CA*R+CB*R+KDA*KDB+KDA*R+KDB*R
114: G1 = INA - KELA*CA - (KINTA*CA*R)/KDA
115: G2 = INB - KELB*CB - (KINTB*CB*R)/KDB
116: G3 = KSYN-KDEG*R-(KINTA*CA*R)/KDA-(KINTB*CB*R)/KDB
117: M11 = (1/DET)*(DET - R*(R+CB+KDB))
118: M12 = (1/DET)*(CA*R)
119: M13 = (1/DET)*(-CA*(R+KDB))
120: M21 = (1/DET)*(CB*R)
121: M22 = (1/DET)*(DET - R*(R+CA+KDA))
122: M23 = (1/DET)*(-CB*(R+KDA))
123: M31 = (1/DET)*(-R*(R+KDB))
124: M32 = (1/DET)*(-R*(R+KDA))
125: M33 = (1/DET)*(DET-CA*(R+KDB)-CB*(R+KDA))
126: DADT(1) = M11*G1 + M12*G2 + M13*G3
127: DADT(2) = M21*G1 + M22*G2 + M23*G3
128: DADT(3) = M31*G1 + M32*G2 + M33*G3
The first lines of the data file are:
150: #ID TIME TYPE DV MDV
151: 1 0 1 . 1
152: 1 0 2 . 1
153: 1 0.0001 1 . 1
154: 1 0.0001 2 . 1
155: 1 2 1 32.9432 0
156: 1 2 2 28.3621 0
ADAPT 5 source code for uncompetitive DDI TMDD
The subroutine DIFFEQ is presented. For full source code see supplemental material.
201: Subroutine DIFFEQ(T,X,XP)
202: Implicit None
203: Include ’globals.inc’
204: Include ’model.inc’
205: Real*8 T,X(MaxNDE),XP(MaxNDE)
206: Real*8 KELA,KDA,KINTA,KELB,KDAB,KINTAB,KSYN,KDEG
207: Real*8 CA,CB,RR,R0
208: Real*8 DET,M(3,3),G(3)
209: KELA = P(1)
210: KDA = P(2)
211: KINTA = P(3)
212: KELB = P(4)
213: KDAB = P(5)
214: KINTAB = P(6)
215: KSYN = P(7)
216: KDEG = P(8)
217: R0 = KSYN/KDEG
218: CA = X(1)
219: CB = X(2)
220: RR = X(3) + R0
221: DET = RR**2*CA+CA*RR*KDA+CB*RR*KDA+CA**2*RR+CA*CB*KDA
222: & +KDA**2*KDAB+KDA*KDAB*RR+CA*KDA*KDAB
223: G(1) = R(1)-KELA*CA-(KINTA*CA*RR)/KDA
224: & -KINTAB*((CA*CB*RR)/(KDA*KDAB))
225: G(2) = R(2)-KELB*CB-KINTAB*((CA*CB*RR)/(KDA*KDAB))
226: G(3) = KSYN-KDEG*RR-(KINTA*CA*RR)/KDA
227: & -KINTAB*((CA*CB*RR)/(KDA*KDAB))
228: M(1,1) = (1/DET)*(DET-RR*(CA*RR+CB*KDA+KDA*KDAB))
229: M(1,2) = (1/DET)*(-CA*RR*KDA)
230: M(1,3) = (1/DET)*(-CA*(CA*RR+CB*KDA+KDA*KDAB))
231: M(2,1) = (1/DET)*(-CB*RR*KDA)
232: M(2,2) = (1/DET)*(DET-CA*RR*(RR+CA+KDA))
233: M(2,3) = (1/DET)*(-KDA*CA*CB)
234: M(3,1) = (1/DET)*(-RR*(CB*KDA+CA*RR+KDA*KDAB))
235: M(3,2) = (1/DET)*(-CA*RR*KDA)
236: M(3,3) = (1/DET)*(DET-CA*(CA*RR+KDAB*KDA+CB*KDA))
237: XP(1) = M(1,1)*G(1)+M(1,2)*G(2)+M(1,3)*G(3)
238: XP(2) = M(2,1)*G(1)+M(2,2)*G(2)+M(2,3)*G(3)
239: XP(3) = M(3,1)*G(1)+M(3,2)*G(2)+M(3,3)*G(3)
240: Return
241: End
Properties of the original DDI TMDD models. Competitive: In panels a and b the single drug profiles of drugs A and B (blue dotted lines) are compared with the competitive model (black solid line) Eqs. (1)–(9) for \(dose_A = 100\), \(dose_B = 100\) and no baseline \(C_A^0 = C_B^0 = 0\). In panels c and d, the effect of the administration of one drug only on a present concentration of the other drug is shown by two examples: (i) drugs A and B are in baseline \(C_A^0 = C_B^0 = 1\), and one administration at time \(t = 12\) of drug B with \(dose_B = 100\) (red dashed lines) causes an increase of drug A concentration, (ii) drug A is administered with \(dose_A = 100\) at \(t = 0\) and drug B administered with \(dose_B = 100\) at \(t = 0\) and additionally at \(t = 12\) (black solid lines), and causes an increase of drug A concentration. Uncompetitive: In panels e and f the single drug profiles of drugs A and B (blue dotted lines) are compared with the uncompetitive model (black solid lines) Eqs. (23)–(33) for \(dose_A = 100\), \(dose_B = 100\) and no baseline \(C_A^0 = C_B^0 = 0\). In panels g and h drugs A and B are in baseline \(C_A^0 = C_B^0 = 1\) with one administration at time \(t=5\) of drug A with \(dose_A = 100\) and non of drug B (red dashed line) and the other way around (black solid lines) (Color figure online)
Visualization of the QE approximation. Competitive: In panels a and b concentration profiles from the original formulation (red dashed lines) Eqs. (1)–(9) and the approximation of the QE formulation Eqs. (14)–(18) (black solid lines) are shown for escalating doses of \(dose_A = dose_B = 10, 100, 1000\) at \(t=0\) and no baseline \(C_A^0 = C_B^0 = 0\). In panels c and d the effect of one drug administration on the present concentration of the other drug is shown. The original (red dashed lines) and QE approximation (black solid lines) profiles with a baseline \(C_A^0 = C_B^0 = 1\) are shown for a dose of \(doseA = 1000\) at \(t=0\). The \(k_{onB}\) and \(k_{offB}\) are multiplied by the factors 0.1, 1 and 10 in such a way that \(K_{DB}\) stays the same to show the convergence of the original formulation towards the QE approximation. Uncompetitive: In panels e and f concentration profiles from the original formulation (red dashed lines) Eqs. (23)–(33) and the approximation of the QE formulation Eqs. (43)–(48) (black solid lines) are shown for escalating doses of \(dose_A = dose_B = 10, 100, 1000\) at \(t=0\) and no baseline \(C_A^0 = C_B^0 = 0\). In panels g and h original (red dashed lines) and QE approximation (black solid lines) profiles with a baseline \(C_A^0 = C_B^0 = 1\) are shown where for drug A is administered with a dose of \(dose_A = 100\) at \(t=24\). The \(k_{onA}\) and \(k_{offA}\) are multiplied by the factors 0.1, 1 and 10 in such a way that \(K_{DA}\) stays the same (Color figure online)
Visualization of plasma concentration versus time data fitting from the original formulation with the QE approximation: Fit (solid lines) of the QE approximation of the competitive Eqs. (14)–(18) in NONMEM (panels a and b) and the uncompetitive DDI TMDD model Eqs. (43)–(48) in ADAPT 5 (panels c and d) in ODE formulation with an IV short infusion. Data (crosses) were produced with the original formulations Eqs. (1)–(9) and Eqs. (23)–(33)
Rights and permissions
About this article
Cite this article
Koch, G., Jusko, W.J. & Schropp, J. Target mediated drug disposition with drug–drug interaction, Part II: competitive and uncompetitive cases. J Pharmacokinet Pharmacodyn 44, 27–42 (2017). https://doi.org/10.1007/s10928-016-9502-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-016-9502-0