Abstract
The immune system is designed to protect an organism from infection and damage caused by a pathogen. A successful immune response requires the coordinated function of multiple cell types and molecules in the innate and adaptive immune systems. Given the complexity of the immune system, it would be advantageous to build computational models to better understand immune responses and develop models to better guide the design of immunotherapies. Often, researchers with strong quantitative backgrounds do not have formal training in immunology. Therefore, the goal of this review article is to provide a brief primer on cellular immunology that is geared for computational modelers.
Similar content being viewed by others
Introduction
The immune system has the daunting task of protecting an organism from disease. It evolved innate and adaptive mechanisms to ward off infection by viruses and bacteria. Additionally, mechanisms exist to eliminate transformed cells arising from an organ, while remaining tolerant to healthy tissues. The extreme variability in antigens, differences between microbes and pathogen life cycles necessitate the need for multiple types of immune cells to mount a productive immune response. Diseases occur when there is a break down in the immune system, such that pathogens evolve to escape an immune response or the immune system attacks host tissues. While much is known about the components of the immune system, it is still difficult to predict if a specific antigen or vaccine candidate will elicit the appropriate immune response needed to prevent or cure a disease. Computational modeling could help provide a more mechanistic understanding of how the immune system functions and yield predictive tools that could help guide the development of immunotherapies. The goal here is to provide modelers a basic primer on cellular immunology.
First contact: what happens when a pathogen enters the body
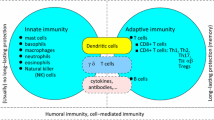
The immune system is constantly scavenging the body to detect infection and disease. Pathogens can enter through penetration of the skin or mucosal epithelium that line the gut, respiratory and urinary tracts. Once inside, pathogens will encounter macrophages and dendritic cells (DC) that reside in tissues, which bear receptors that recognize specific components of a pathogen (Fig. 1). These receptors include Toll-like receptors (TLR) [1], scavenger receptors [2], and mannose receptors [3]. Most of these are cell surface receptors. However, TLR-3, 7, and 9 reside in the endosome and recognize dsRNA, ssRNA and unmethylated CpG DNA respectively [4–6]. Many pathogens are engulfed and internalized in a phagosomal vesicle, which, following fusion with the lysosome, will lead to the destruction of the pathogen. The interaction with the pathogen also stimulates macrophages and DC to release cytokines and chemokines including IL-1, IL-6, IL-8, IL-12 and TNF-α [7–9]. Cytokine and chemokine release initiate inflammatory pathways and recruit other immune cells to the site of infection, including neutrophils and monocytes [10]. Neutrophils destroy pathogens through the release of reactive oxygen and nitrogen species and lysosomal degradation. DC that interact with pathogens in the tissues are induced to undergo maturation [11]. Mature DCs change their pattern of chemokine receptor expression and begin to migrate towards the lymph node (LN) that drains the initial infection site [12]. In addition, DC upregulate expression of molecules necessary for the presentation of pathogen-derived antigens to the adaptive immune system. The initial phase of an infection in part involves pathogen recognition, cytokine production, and recruitment of appropriate immune cells.
Activation and function of macrophages. Macrophages express pattern recognition receptors on the cell surface. These include the mannose receptor, Toll receptors such as TLR-4, and scavenger receptor. Upon binding a pathogen, the macrophage will produce and secrete lipids, cytokines and chemokines that have effector functions to promote an immune response. Additionally, the macrophage will engulf the pathogen. A lysosome will fuse with an endosome containing the pathogen. Enzymes from the lysosomes will degrade the pathogen
Inflammatory responses that are initiated by macrophage and DC cytokine secretion early in an infection are important for promoting an immune response. Inflammation alters the site of infection. Blood vessels proximal to the infection dilate allowing for increased local blood flow and increased delivery of immune cells. Activated endothelial cells along the blood vessel up regulate the expression of cell adhesion proteins resulting in leukocytes adhering to the blood vessel wall [13]. After adhesion, leukocytes then migrate into the infected tissue. The first cells to migrate to a site of infection are neutrophils, which can directly kill extracellular pathogens [14, 15]. Next, monocytes arrive, which can differentiate into either tissue specific macrophages or DC. The macrophages can directly eliminate a pathogen and can also present antigens to the adaptive immune system. Activated macrophages and DC secrete chemokines and cytokines to recruit leukocytes like eosinophils and lymphocytes [16]. For viral infections, infected cells secrete IFNs that inhibit viral replication and activate natural killer cells to directly destroy infected cells [17]. A critical step in the early response is the migration of mature DC to draining LNs where the adaptive immune response is initiated.
Antigen processing and presentation on cell surfaces
DCs are specialized antigen presenting cells (APC) that are uniquely able to activate naïve T cells [12]. The activation of T cells requires recognition of pathogen-derived antigens bound in the groove of major histocompatibility complex (MHC) molecules present of the surface of APC [18, 19]. The two forms of MHC molecules, MHC class I (MHC I) and MHC class II (MHC II), sample antigens from different cell compartments. In general, MHC II molecules bind peptides derived from exogenously-derived proteins whereas MHC I bind peptides derived from cytoplasmic sources. MHC I molecules are expressed on all cells of the body whereas MHC II is only expressed by professional APC (DC, macrophages, B cells) or by tissue cells in inflammatory conditions following exposure to interferon (IFN).
Bacteria, bacterial antigens or other pathogens in the extracellular space can be encapsulated into intracellular vesicles called endosomes by macrophages, immature DC, and many other cell types [1] (Fig. 2a). Alternatively, pathogens can replicate in endosomes of a cell. The internalized endosomes have a neutral pH and contain proteases that are inactive. Acidification of the endosome activates the endosomal proteases, which then digest the antigens into small peptide fragments [20]. Vesicles containing MHC II then fuse with vesicles containing the digested antigenic peptides. Initially MHC II is complexed with an invariant chain [21]. Upon fusion with the acidic endosome, the invariant chain is cleaved by the activated proteases and a short peptide fragment, known as CLIP, remains bound to MHC II. The HLA-DM molecule, which is an MHC II-like protein, catalyzes the release of the CLIP peptide from MHC II and allows the antigenic peptide fragments to be loaded into MHC II. The peptide loaded MHC II then traffics to the cell surface where it can bind to the T cell receptor (TCR) expressed on CD4+ T cells [22].
Antigen processing and MHC antigen presentation. a Cells such as macrophages or dentric cells will endocytose an antigen. These cells also produce the MHC II protein, which is comprised of an α and β chain. Initially, MHC II is assembled in the ER and associates with the invariant chain in a specialized MIIC compartment. The invariant chain blocks peptides from entering the MHC II peptide-binding cleft. The endosome containing the MHC II and the endosome containing the antigen will acidify, which triggers the activation of proteases that will degrade the invariant chain and the antigen. The endosomes will fuse and HLA-DM will exchange the invariant peptide for an antigenic peptide. The MHC II loaded with the antigenic peptide will then traffic to and be presented on the cell surface where it can be surveyed by T cells. b Some pathogens including viruses hijack the host cell to synthesize proteins. When a viral protein is synthesized, it can be UB and degraded into small peptide fragments by the proteasome. These proteins can be imported into the ER by TAP. The MHC I molecule remains in a partially folded state in the ER. Binding the antigenic peptide will result in the proper folding of MHC I. The folded peptide-MHC I will traffic to and be presented on the cell surface where it can interact with receptors on cytotoxic T cells
Some viruses and bacteria replicate in the cytosol or nuclear compartment. After pathogenic proteins are synthesized, they can be ubiquitinated (UB) and degraded by the proteasome [23] (Fig. 2b). These peptide antigens are transported from the cytoplasm to the lumen of the endoplasmic reticulum (ER) by the TAP transporter [24]. The newly synthesized MHC I resides in the lumen of the ER. Initially, MHC I is in a partially folded state that binds to β2 microglobulin. Antigenic peptides transported by TAP can then bind to MHC I. Once a stable peptide-MHC I complex is formed, it is trafficked to and presented on the cell surface of the infected cell, where it can interact with the TCR on a CD8+ T cell [25].
T cell mediated immune responses
Structural organization of TCRs
T cells recognize peptide MHC (pMHC) complexes present on the surfaces of APC including DC, macrophages, B cells and diseased cells using their associated TCR. TCRs are heterodimeric proteins comprised of an α and β chain (Fig. 3a). Each chain has a variable and constant domain. The constant domains interact with the CD3 complement of co-receptors that are involved with signaling [26]. The variable domain contains complementary determining loops (CDR) that bind to pMHCs. The CDR loops are generated via somatic recombination [27], which produces TCRs with many different antigen specificities. As described above antigenic peptide are presented by class I and class II MHCs. MHC I is composed of the MHC I α chain, β2 microglobulin (β2m) and antigenic peptide [28] (Fig. 3b). MHC II is composed of the antigenic peptide, β chain and α chain [29] (Fig. 3c). For both MHC I and MHC II presented antigens, the αβ TCR binds to the composite surface formed by the antigenic peptide and MHC [30, 31] (Fig. 3d and e). Recent biophysical studies demonstrate that the pMHC I is a highly dynamic molecule. The antigenic peptide can adopt multiple conformations and the α1 and α2 helices of MHC I are also mobile [32, 33]. To deal with this complexity, the CDR loops of the TCR are flexible and are able to meld to the pMHC to form a stable complex [34, 35].
TCR recognition of antigens presented by MHC I and MHC II a TCR are transmembrane proteins expressed on the surface of T cells. TCRs are heterodimers comprised of an α and β chain (Protein Data Bank (PDB) ID 1TCR). Each chain contains a constant, C, and variable, V, domain. The constant domains associate with coreceptors on the T cell, while the variable domain functions to bind peptide-MHC. b The MHC I is a trimolecular complex comprised of the MHC chain, β2 microglobulin, and the antigenic peptide (PDB ID 1BD2). c The MHC II complex is comprised of a peptide and α and β chain. The TCRs bind to both the antigenic peptide and MHC I (PDB ID 1BD2) d and peptide-MHC II surfaces (PDB ID 3T0C) e T cells express coreceptors that are important for signaling. For example, Th cells express CD4 on their cell surface. f The CD4 molecule binds to a different region of the MHC II than the TCR (PDB ID 3T0C)
In addition to TCRs, T cells express co-receptors that bind to molecules on APC and serve to increase the avidity of the interaction. CD4+ T cells (also known as “helper cells”) express the CD4 protein on the cell surface. Upon ligation of the TCR to a cognate MHC II, the CD4 protein will also bind to the MHC II molecule [36] (Fig. 3f). Likewise, CD8+ T cells (also known as “killer cells”) express the CD8 protein. After a TCR interacts with MHC I, CD8 will also bind to the MHC I molecule. Recent work demonstrates that TCR binding rigidifies the CD8 binding site on a MHC I molecule [32]. This could facilitate CD8 coreceptor binding. In addition to molecules such as CD4 and CD8 that bind MHC molecules, T cells express costimulatory molecules including CD28, which recognizes CD80 and CD86 that are expressed on DC and other APC [37]. Together, TCR ligation to MHC and activation of costimulatory molecules initiates T cell activation.
Activation of T cells
Following stable formation of the TCR-MHC complex, members of the SRC family of protein threonine kinases such as LCK are activated. LCK phosphorylates intracellular segments of CD3 proteins in the TCR complex including the CD3 ζ heterodimer. Phosphorylation of ζ recruits ZAP70, a member of the SYK family of protein tyrosine kinases [38, 39]. ZAP70 then phosphorylates multiple targets including SLP-76 and LAT [40], which recruit other proteins including phospholipase C. Downstream signaling events including calcium release, reorganization of the actin cytoskeleton and activation of transcription factors leading to activation of T cells. Binding of costimulatory molecules including CD28 are necessary to amplify the signal and to induce optimal T cell activation [37]. Recognition of pMHC in the absence of CD28 engagement leads to T cell anergy and is one mechanism for the maintenance of tolerance to self tissues [41]. TCR signaling is also termed signal 1. Binding CD80 or CD86 activates signaling through CD28 and constitutes signal 2. Signal 1 and signal 2 promote T cell proliferation through secretion of IL-2 and expression of CD25, which is the high affinity component of the IL-2 receptor.
Differentiation and effector functions of T cells
Following the initial recognition of antigen through signal 1 and signal 2 CD4 T cells proliferate and differentiate into various effector cell populations. These T helper (Th) subsets are characterized by a unique pattern of cytokine expression, which is controlled by specific master transcription factors [42] (Fig. 4a). Th subsets have evolved to deal with specific pathogen threats. Th cells can differentiate into Th1, Th2, Th17, Treg or Tfh cells (Fig. 4a) and the list is growing. Th2 cells are involved with the immune response against helminth worms and secrete IL-4, IL-5 and IL-13, which activates basophils, eosinophils and mast cells. Th1 cells are important for defense against intracellular pathogens and viruses and the major cytokine secreted by Th1 cells is IFN-γ, which stimulates macrophages, CD8+ T cells and induces isotype switching in B cells. In addition to Th1 and Th2 cells, CD4+ T cells can differentiate into Tfh, Th17 and Treg subtypes, which are important in germinal centers, defense against extracellular pathogens and immunosuppression respectively.
Effector functions of T cells a CD4+ T cells can differentiate into multiple subsets of cells. The types of cytokines and other environmental factors drive this cell fate decision. Each subtype utilizes different transcription factors. For example, induction of Treg development is associated with Foxp3 expression. Each subset expresses a different complement of cytokines, which promotes different types of immune responses. b The major function of CD8+ T cells is to destroy infected or transformed cells. Binding of the TCR to its cognate peptide-MHC I ligand promotes T cell activation and subsequent secretion of perforin and granzymes. Perforin will form a pore structure in the infected cell. The perforin pore allows granzymes and other compounds secreted to enter the target cell. Granzymes are proteases and activate the caspase cascade that results in apoptosis and subsequent death of the target cell
Immediately after activation CD4 T cells (known as Th0) secrete IL-2, and low levels of IL-4 and IFN-γ [43]. T cells differentiate into specific Th subsets based on the cytokines produced by the DC that presents the antigen. This cytokine signal is known as signal 3. DC secrete different cytokines depending on the type of pathogen it encountered [44]. The types of cytokines secreted by the DC drive the differentiation of the Th cell subset; IL-12 stimulates IFN-γ production and Th1 differentiation whereas IL-23, IL-6 and TGF-β are important for Th17 differentiation [45]. Other factors that are important for Th cell differentiation include the dose of the stimulating antigen and specific costimulatory molecules.
Th cells function to “help” other immune cells by secreting cytokines to orchestrate an immune response. Th cells function to stimulate antibody production by B cells, activate cytotoxic T cells, increase macrophage function and modulate immune responses to prevent autoimmunity. After the T cell encounters a DC in the LN, effector and memory cells will develop. At this stage, differentiated Th cells also acquire chemokine receptors that allow Th cells to leave the LN and migrate to the site of the infection. Once effector T cells migrate to the site of infection and encounter a cell expressing a cognate pMHC molecule they secrete the appropriate cytokines to enable the destruction of the pathogen [42]. For example, in the case of an infection with an intracellular pathogen Th1 cells secrete IFN-γ, which activates tissue macrophages to produce reactive oxygen and nitrogen species and increases MHC and costimulatory molecule expression levels leading to more efficient killing and antigen presentation by macrophages. In the case of infection with extracellular bacteria Th17 cells secrete IL-17, which acts on local tissues to increase production of IL-8, a potent neutrophil chemoattractant. Memory cells are also generated and these are long-lived cells, which circulate both in tissues and LNs where they can give rise to a more rapid response upon a second encounter with the same pathogen.
Cytotoxic T cells express CD8 on their cell surface, and thus respond to APC that bear MHC I ligands (Fig. 4a). CD8+ T cells mediate immune responses to cancer cells and virally infected cells. The major function of CD8+ T cells is to directly destroy diseased cells [46]. When a CD8+ T cell is stimulated in a LN by a DC expressing a cognate pMHC I ligand, the T cell proliferates and generates effector and memory T cells. Optimal CD8+ T cell activation and differentiation also requires cytokines from Th1 cells. The CD8+ T cell migrates to the site of infection. When the TCR on the CD8+ T cell binds to a virally infected cell with a cognate pMHC I ligand, the effector CD8+ T cell releases its cytotoxic granules that contain perforin and granzymes. Perforin forms a pore in the target cell that facilitates granzyme entry, which will then initiate a caspase cascade that triggers apoptosis in the targeted cell [47] (Fig. 4b).
B cell mediated effector functions
A major function of B cells is to secrete antibodies to mediate a humoral immune response. For a B cell to successfully produce and secrete an antibody, multiple steps need to occur (Fig. 5). Each B cell expresses a unique B cell receptor (BCR) on the cell surface that recognizes a cognate antigen. The structure of the BCR is very similar to an antibody except that it is a transmembrane protein and is associated with two immunoreceptor tyrosine-based activation motif (ITAM) containing signaling chains, Igα and Igβ. Naïve B cells are present within the follicles of LNs and spleen and encounter antigen that drains into the LN from a site of infection or interact with DC and macrophages that present the antigen. After a BCR binds antigen, the antigen is internalized, degraded into peptide fragments and MHC II presents the peptide antigens on the B cell surface [48]. A cognate TCR on a CD4+ T cells can then bind to the pMHC II molecule. The T cell becomes activated and produces the CD40L, which will bind to the CD40 receptor [49]. Additionally, the helper T cell will produce IL-4. These signals from the Th cell along with direct signaling from the BCR activate the B cell to proliferate and undergo class switching (see below). The initial interaction between B and T cells occurs in the region between the follicle and T cell area of the LN.
B cell activation and antibody production B cells express the BCR and present antigens via MHC II. The BCR is akin to a membrane bound antibody. Antigen binding results in BCR cross-linking and signals activation of the B cell. A Th cell binds to the pMHC II presented on the B cell via its TCR. Activation of the T cells induces expression of CD40L, which binds to CD40 on the B cell. These processes lead to cytokine secretion from the T cell and subsequent activation of the B cell. Through a complicated process known as affinity maturation the BCR will undergo rounds of hypermutation to enhance its affinity for the antigen. The B cell proliferates and differentiates intomemory B cells and effector cells called plasma cells. The plasma cells secrete antibodies towards the antigen
B cells undergo several rounds of differentiation to become plasma cells that secrete antibodies [50]. Some of the proliferating B cells differentiate into plasmablasts that are capable of producing and secreting antibodies. Plasmablasts actively divide and retain hallmarks of activated B cells that allow plasmablasts to interact with T cells. Some of the plasmablasts will further differentiate into plasma cells, while others will die. Plasma cells lose surface expression of MHC and express lower levels of the BCR. Many of the generated plasma cells migrate to the bone marrow and continue secreting antibodies. Some activated B cells migrate into the follicle, along with specialized Tfh cells, and form a germinal center [51]. In the germinal center, B cells undergo a process called affinity maturation [52]. During affinity maturation, mutations of the V regions of the immunoglobulin gene occur. Mutations that result in a BCR with lower antigen affinity undergo apoptosis and die, while mutations that result in a BCR with high antigen affinity differentiate into either memory B cells or plasma cells (Fig. 5). The B cell tests the affinity of the mutated antibody through interactions with antigen presented by follicular DC and by the ability to present the antigen to Tfh cells within the germinal center
Mechanisms of antibody mediated immunity
The variable domains of an antibody determine the antigen specificity, while the heavy chain constant domains determine the effector function. The major classes of antibodies in humans are IgA, IgD, IgE, IgG, and IgM. Naïve B cells first secrete IgM, and IgM therefore mediates the early antibody response. At later stages, IgM only constitutes ten percent of the total antibody pool, and the IgG and IgA isotypes become dominant. The change in antibody isotype generated by a B cell is called class switching [53]. Cytokines produced by Th cells direct class switching and determine which antibody isotype a B cell will make.
Each antibody isotype has specialized effector functions including neutralization of a pathogen and recruitment of immune cells and activation of effector cells (Fig. 6a). For example, IgM forms pentamers and bind to the surface of bacteria [54] (Fig. 6b). The bound IgM molecule then activates the complement cascade, which results in the destruction of the bacteria. IgG antibodies coat a pathogen resulting in uptake and destruction by a phagocytic cell [55]. Many cells in the immune system have receptors that recognize antibody-coated pathogens or cells. For example, macrophages express Fc receptors that bind to aggregated antibodies on the surface of a bacteria. There is an Fc receptor specific for each of the isotypes and there are six different Fc receptors that bind IgG isotypes. In general these Fc receptors are activating receptors since they also associated with ITAM containing signaling chains. The cross-linked Fc receptor activates the macrophage, which results in the phagocytosis and ultimate destruction of the bacteria (Fig. 6c). NK cells are cytotoxic lymphocytes that express Fc receptors specific for IgG (Fig. 6d). Virally infected cells often express viral proteins on the surface of the infected cell. Antibodies recognize these antigens. Fc receptors on NK cells bind to the antibodies on the surface of the virally infected cell and the cross-linked Fc receptors stimulate the NK cell to degranulate releasing perforin and granzymes into the space between the cells [56]. This results in apoptosis of the infected cell. Thus Fc receptors provide innate immune cells with the specificity of the adaptive immune system through the binding of antigen-specific antibody [57]. IgA antibodies are secreted into the lumen of mucosal tissues and provide a barrier function in the gut and lung [58]. IgE is induced by Th2 cells and is a critical mediator of allergic reactions [59]. IgE binds to a high affinity Fc receptor on mast cells and crosslinking of this receptor with an allergen causes mast cell degranulation with release of histamine and other mediators of the allergic response. These examples highlight the diversity and importance of antibody mediated immune responses.
Some functions of antibodies. Antibodies have multiple functions. a Some antibodies bind to receptors on viruses or bacteria, which blocks or “neutralizes” the virus from interacting with the host cell. b IgM antibodies form pentamers that bind to antigens on the surfaces of bacteria or cells. The antibodies target complement to the bacteria and activate the complement pathway to destroy the pathogen. c Many phagocytic cells including macrophages express Fc receptors. Fc receptors bind to antibodies. When a pathogen is coated with antibodies, the targeted pathogen can be phagocytosed and destroyed by macrophages. d NK cells also express Fc receptors. When a cell is infected with a virus, viral proteins are often expressed on the cell surface of the host. Antibodies bind to these surface antigens. The Fc receptors on the NK cell recognize these aggregated antibodies on the surface of the infected cell resulting in NK activation, release of cytotoxic granules and death of the infected cell
Control of the immune response
Once a pathogen has been destroyed it is necessary for the system to return to homeostasis and for the damaged tissue to be repaired. There are multiples levels of control of the immunes response and these are necessary to prevent an over zealous immune response that causes significant tissue damage. One such mechanism is the secretion of the cytokine IL-10. IL-10 is produced by many immune cells and its major function is to inhibit the activation of macrophages and other effector cells. When IL-10 is deleted in mouse systems, mice will die following infection as a result of excessive inflammatory cytokine production, despite the fact that the pathogen was successfully cleared. Treg cells also play am important role in controlling ongoing immune responses through the secretion of cytokines such as IL-10 and TGF-β [60]. Treg also play a critical role in preventing autoimmunity in which cells of the immune system may inappropriately respond to self-antigens and induce tissue destruction. Control of antibody responses is mediated in part by inhibitory Fc receptors. B cells and macrophages express the inhibitory Fc gamma receptor that binds IgG. This functions to prevent the generation of auto-antibodies by B cells and also to limit the function and activation of macrophages.
Conclusion
The function of the immune system is to prevent infection and disease. Given the large universe of potential pathogens and diversity in pathogen life cycles, the immune system has evolved multiple mechanisms to eliminate pathogens and diseased cells. This is accomplished through the coordinated effort of different types of cells and effector molecules. Innate immune cells such as macrophages and neutrophils act early to contain and possibly eliminate a pathogen. If a pathogen escapes innate immune responses, then cells of the adaptive immune system that more specifically target a pathogen can respond to eliminate an infection. The response initiated by each type of cell in the immune cell is driven by intracellular signaling events, the cytokine environment in which it is stimulated and physical interactions with cells and tissues. The specificity of the immune response has stimulated much work focused on harnessing the power of the immune system for immunotherapies, including for diseases such as cancer, autoimmune disease and vaccine development. This requires detailed models of the immune system built at multiple scales that span intracellular biochemical signaling pathways, cellular interactions within tissues to a broader appreciation of the how immune system cells function in an entire organism. Computational modeling approaches have the potential to generate predictive models that will provide a more integrated view of the immune system.
References
Aderem A, Underhill DM (1999) Mechanisms of phagocytosis in macrophages. Annu Rev Immunol 17:593–623
Gough PJ, Gordon S (2000) The role of scavenger receptors in the innate immune system. Microbes Infect 2(3):305–311
Apostolopoulos V, McKenzie IF (2001) Role of the mannose receptor in the immune response. Curr Mol Med 1(4):469–474
Barton GM, Medzhitov R (2002) Toll-like receptors and their ligands. Curr Top Microbiol Immunol 270:81–92
Lund J et al (2003) Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med 198(3):513–520
Lund JM et al (2004) Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA 101(15):5598–5603
Larsson BM et al (1999) Gram positive bacteria induce IL-6 and IL-8 production in human alveolar macrophages and epithelial cells. Inflammation 23(3):217–230
Svanborg C, Godaly G, Hedlund M (1999) Cytokine responses during mucosal infections: role in disease pathogenesis and host defence. Curr Opin Microbiol 2(1):99–105
Ono SJ et al (2003) Chemokines: roles in leukocyte development, trafficking, and effector function. J Allergy Clin Immunol 111(6):1185–1199; quiz 1200
Kunkel EJ, Butcher EC (2002) Chemokines and the tissue-specific migration of lymphocytes. Immunity 16(1):1–4
Ardavin C (2003) Origin, precursors and differentiation of mouse dendritic cells. Nat Rev Immunol 3(7):582–590
Gatti E, Pierre P (2003) Understanding the cell biology of antigen presentation: the dendritic cell contribution. Curr Opin Cell Biol 15(4):468–473
Alon R, Feigelson S (2002) From rolling to arrest on blood vessels: leukocyte tap dancing on endothelial integrin ligands and chemokines at sub-second contacts. Semin Immunol 14(2):93–104
Scapini P et al (2000) The neutrophil as a cellular source of chemokines. Immunol Rev 177:195–203
Bochenska-Marciniak M et al (2003) The effect of recombinant interleukin-8 on eosinophils’ and neutrophils’ migration in vivo and in vitro. Allergy 58(8):795–801
Yoshie O (2000) Role of chemokines in trafficking of lymphocytes and dendritic cells. Int J Hematol 72(4):399–407
Kawai T, Akira S (2006) Innate immune recognition of viral infection. Nat Immunol 7(2):131–137
Villadangos JA (2001) Presentation of antigens by MHC class II molecules: getting the most out of them. Mol Immunol 38(5):329–346
Gromme M, Neefjes J (2002) Antigen degradation or presentation by MHC class I molecules via classical and non-classical pathways. Mol Immunol 39(3–4):181–202
Lennon-Dumenil AM et al (2002) A closer look at proteolysis and MHC-class-II-restricted antigen presentation. Curr Opin Immunol 14(1):15–21
Gregers TF et al (2003) The cytoplasmic tail of invariant chain modulates antigen processing and presentation. Eur J Immunol 33(2):277–286
Pathak SS, Lich JD, Blum JS (2001) Cutting edge: editing of recycling class II:peptide complexes by HLA-DM. J Immunol 167(2):632–635
Goldberg AL et al (2002) The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol Immunol 39(3–4):147–164
Gorbulev S, Abele R, Tampe R (2001) Allosteric crosstalk between peptide-binding, transport, and ATP hydrolysis of the ABC transporter TAP. Proc Natl Acad Sci USA 98(7):3732–3737
Ackerman AL, Cresswell P (2004) Cellular mechanisms governing cross-presentation of exogenous antigens. Nat Immunol 5(7):678–684
Ghendler Y et al (1998) One of the CD3epsilon subunits within a T cell receptor complex lies in close proximity to the Cbeta FG loop. J Exp Med 187(9):1529–1536
Shinkai Y et al (1992) RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68(5):855–867
Bjorkman PJ et al (1987) Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329(6139):506–512
Fremont DH et al (1992) Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science 257(5072):919–927
Garcia KC et al (1996) An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 274(5285):209–219
Ding YH et al (1998) Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity 8(4):403–411
Hawse WF et al (2012) Cutting edge: evidence for a dynamically driven T cell signaling mechanism. J Immunol 188(12):5819–5823
Hawse WF et al (2013) Peptide modulation of class I major histocompatibility complex protein molecular flexibility and the implications for immune recognition. J Biol Chem 288(34):24372–24381
Hawse WF et al (2014) TCR scanning of peptide/MHC through complementary matching of receptor and ligand molecular flexibility. J Immunol 192(6):2885–2891
Baker BM et al (2012) Structural and dynamic control of T-cell receptor specificity, cross-reactivity, and binding mechanism. Immunol Rev 250(1):10–31
Li Y, Yin Y, Mariuzza RA (2013) Structural and biophysical insights into the role of CD4 and CD8 in T cell activation. Front Immunol 4:206
Appleman LJ et al (2000) CD28 costimulation mediates T cell expansion via IL-2-independent and IL-2-dependent regulation of cell cycle progression. J Immunol 164(1):144–151
Chan AC et al (1992) ZAP-70: a 70 kd protein-tyrosine kinase that associates with the TCR zeta chain. Cell 71(4):649–662
Chan AC et al (1995) Activation of ZAP-70 kinase activity by phosphorylation of tyrosine 493 is required for lymphocyte antigen receptor function. EMBO J 14(11):2499–2508
Jordan MS, Singer AL, Koretzky GA (2003) Adaptors as central mediators of signal transduction in immune cells. Nat Immunol 4(2):110–116
Harding FA et al (1992) CD28-mediated signalling co-stimulates murine T cells and prevents induction of anergy in T-cell clones. Nature 356(6370):607–609
Geginat J et al (2013) The CD4-centered universe of human T cell subsets. Semin Immunol 25(4):252–262
Murphy KM, Reiner SL (2002) The lineage decisions of helper T cells. Nat Rev Immunol 2(12):933–944
Reis e Sousa C et al (1997) In vivo microbial stimulation induces rapid CD40 ligand-independent production of interleukin 12 by dendritic cells and their redistribution to T cell areas. J Exp Med 186(11):1819–1829
Manel N, Unutmaz D, Littman DR (2008) The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol 9(6):641–649
Zhang N, Bevan MJ (2011) CD8(+) T cells: foot soldiers of the immune system. Immunity 35(2):161–168
Thiery J et al (2011) Perforin pores in the endosomal membrane trigger the release of endocytosed granzyme B into the cytosol of target cells. Nat Immunol 12(8):770–777
Lanzavecchia A (1990) Receptor-mediated antigen uptake and its effect on antigen presentation to class II-restricted T lymphocytes. Annu Rev Immunol 8:773–793
Jaiswal AI, Croft M (1997) CD40 ligand induction on T cell subsets by peptide-presenting B cells: implications for development of the primary T and B cell response. J Immunol 159(5):2282–2291
Shapiro-Shelef M, Calame K (2005) Regulation of plasma-cell development. Nat Rev Immunol 5(3):230–242
Kelsoe G (1996) The germinal center: a crucible for lymphocyte selection. Semin Immunol 8(3):179–184
Ziegner M, Steinhauser G, Berek C (1994) Development of antibody diversity in single germinal centers: selective expansion of high-affinity variants. Eur J Immunol 24(10):2393–2400
Stavnezer J (1996) Immunoglobulin class switching. Curr Opin Immunol 8(2):199–205
Koshland ME (1985) The coming of age of the immunoglobulin J chain. Annu Rev Immunol 3:425–453
Clark MR (1997) IgG effector mechanisms. Chem Immunol 65:88–110
Sulica A et al (2001) Ig-binding receptors on human NK cells as effector and regulatory surface molecules. Int Rev Immunol 20(3–4):371–414
Ward ES, Ghetie V (1995) The effector functions of immunoglobulins: implications for therapy. Ther Immunol 2(2):77–94
Brandtzaeg P (2003) Role of secretory antibodies in the defence against infections. Int J Med Microbiol 293(1):3–15
Galli SJ, Tsai M (2012) IgE and mast cells in allergic disease. Nat Med 18(5):693–704
Takahashi T, Sakaguchi S (2003) Naturally arising CD25+ CD4+ regulatory T cells in maintaining immunologic self-tolerance and preventing autoimmune disease. Curr Mol Med 3(8):693–706
Acknowledgments
We greatly acknowledge grant support from the US National Institutes of Health (5T32AI089443-04).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hawse, W.F., Morel, P.A. An immunology primer for computational modelers. J Pharmacokinet Pharmacodyn 41, 389–399 (2014). https://doi.org/10.1007/s10928-014-9384-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-014-9384-y