Abstract
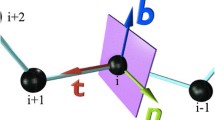
A protein is commonly visualized as a discrete piecewise linear curve, conventionally characterized in terms of the extrinsically determined Ramachandran angles. However, in addition to the extrinsic geometry, the protein has also two independent intrinsic geometric structures, determined by the peptide planes and the side chains respectively. Here we develop a novel 3D visualization method that instead of the extrinsic geometry utilizes the intrinsic geometry of side chains. We base our approach on a series of orthonormal coordinate frames along the protein side chains in combination with a mapping of the atoms positions onto a unit sphere, for visualization purposes. We develop our methodology in terms of an example, by analyzing the acidity dependence of the presumed myoglobin ligand gate. In the literature, the ligand gate is often asserted to be highly localized at the distal histidine, its functioning being regulated by environmental changes. Thus, we investigate whether any \({\mathrm{pH}}\) dependence can be detected in the orientation of the distal and proximal histidine residues, using existing crystallographic data. We observe no \({\mathrm{pH}}\) dependence, in support of the alternative proposals that the ligand gate is more complex and might even be located elsewhere. Our methodology should help the planning of future myoglobin structure experiments, to identify the ligand gate position and its mechanism. More generally, our methodology is designed to visually depict the spatial orientation of side chain covalent bonds in a protein. As such, it can be eventually advanced into a general visual 3D tool for protein structure analysis for purposes of prediction, validation and refinement. It can serve as a complement to widely used visualization suites such as VMD, Jmol, PyMOL and others.








Similar content being viewed by others
References
https://en.wikipedia.org/wiki/List of molecular graphics systems. Accessed 14 Sept 2018
H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne, The protein data bank. Nucl. Acids Res. 28(1), 235–242 (2000)
P.A. Calligari, G.R. Kneller, ScrewFit: combining localization and description of protein secondary structure. Acta Crystallogr. Sect. D Biol. Crystallogr. 68(12), 1690–1693 (2012)
D. Case, M. Karplus, Dynamics of ligand binding to heme proteins. J. Mol. Biol. 132(3), 343–368 (1979)
P.S. Coelho, E.M. Brustad, A. Kannan, F.H. Arnold, Olefin cyclopropanation via carbene transfer catalyzed by engineered cytochrome p450 enzymes. Science 339, 307–310 (2013). http://science.sciencemag.org/content/339/6117
J. Du, M. Sono, J.H. Dawson, The H93G myoglobin cavity mutant as a versatile scaffold for modeling heme iron coordination structures in protein active sites and their characterization with magnetic circular dichroism spectroscopy. Coord. Chem. Rev. 255(7–8), 700–716 (2011)
R. Elber, M. Karplus, Enhanced sampling in molecular dynamics: use of the time-dependent hartree approximation for a simulation of carbon monoxide diffusion through myoglobin. J. Am. Chem. Soc. 112(25), 9161–9175 (1990)
H. Frauenfelder, B.H. McMahon, P.W. Fenimor, Myoglobin: the hydrogen atom of biology and a paradigm of complexity. Proc. Natl. Acad. Sci. USA 100, 8615–8617 (2003)
Y. Hou, J. Dai, J. He, A. Niemi, X. Peng, N. Ilieva, Intrinsic protein geometry with application to non-proline cis peptide planes. J. Math. Chem. 57(1), 263 (2019)
X. Huang, S.G. Boxer, Discovery of new ligand binding pathways in myoglobin by random mutagenesis. Nat. Struct. Biol. 1(4), 226–229 (1994)
J.C. Kendrew, G. Bodo, H.M. Dintzis, R.G. Parrish, H. Wyckoff, D.C. Phillips, Three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature 181, 662–666 (1958)
J .C. Kendrew, R.E. ans Dickerson, B.E. Strandberg, R.G. Hart, D.R. Davies, D.C. Phillips, V.C. Shore, Structure of myoglobin: a three-dimensional fourier synthesis at 2 Å resolution. Nature 185, 422–427 (1960)
G.R. Kneller, K. Hinsen, Protein secondary-structure description with a coarse-grained model. Acta Crystallogr. Sect. D Biol. Crystallogr. 71(7), 1411–1422 (2015)
A. Krokhotin, A. Niemi, X. Peng, On the role of thermal backbone fluctuations in myoglobin ligand gate dynamicsy. J. Chem. Phys. 138(17), 175101 (2013)
Y. Lin, J. Wang, Y. Lu, Functional tuning and expanding of myoglobin by rational protein design. Sci. China Chem. 57(3), 346–355 (2014)
G. Maggiora, P. Mezey, B. Mao, K. Chou, A new chiral feature in \(\alpha \)-helical domains of proteins. Biopolym. Orig. Res. Biomol. 30(1–2), 211–214 (1990)
P.G. Mezey, K. Fukui, S. Arimoto, Treatment of small deformations of polyhedral shapes of functional group distributions in biomolecules. Int. J. Quantum Chem. 76(6), 756–761 (2000)
P.G. Mezey, K. Fukui, S. Arimoto, K. Taylor, Polyhedral shapes of functional group distributions in biomolecules and related similarity measures. Int. J. Quantum Chem. 66(1), 99–105 (1998)
S. Mondal, S. Ghosh, Effect of curcumin on the binding of cationic, anionic and nonionic surfactants with myoglobin. J. Mol. Struct. 1134, 292–297 (2017)
J.S. Olson, A.J. Mathews, R.J. Rohlfs, B.A. Springer, K.D. Egeberg, S.G. Sligar, J. Tame, J.P. Renaud, K. Nagai, The role of the distal histidine in myoglobin and haemoglobin. Nature 336(6196), 265 (1988)
X. Peng, A. Chenani, S. Hu, Y. Zhou, A.J. Niemi, A three dimensional visualisation approach to protein heavy-atom structure reconstruction. BMC Struct. Biol. 14(1), 27 (2014)
M.F. Perutz, F. Mathews, An X-ray study of azide methaemoglobin. J. Mol. Biol. 21(1), 199–202 (1966)
M.F. Perutz, M.G. Rossman, A.F. Cullis, H. Muirhead, G. Will, A.C.T. North, Structure of haemoglobin: a three-dimensional fourier synthesis at 5.5-Å. resolution, obtained by X-ray analysis. Nature 185, 416–422 (1960)
S.E. Phillips, B.P. Schoenborn, Neutron diffraction reveals oxygen-histidine hydrogen bond in oxymyoglobin. Nature 292(5818), 81 (1981)
G. Ramachandran, C. Ramakrishnan, V. Sasisekharan, Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 7, 95–99 (1963)
I. Schlichting, J. Berendzen, G.N. Phillips Jr., R.M. Sweet, Crystal structure of photolysed carbonmonoxy-myoglobin. Nature 371, 808–812 (1994)
M.D. Spivak, A Comprehensive Introduction to Differential Geometry (Publish or Perish, Berkeley, 1970)
T.Y. Teng, W. Schildkamp, P. Dolmer, K. Moffat, Two open-flow cryostats for macromolecular crystallography. J. Appl. Crystallogr. 27(2), 133–139 (1994)
R. Tilton, I. Kuntz, G. Petsko, Cavities in proteins: structure of a metmyoglobin-xenon complex solved to 1.9 Å. Biochemistry 23, 2849–2857 (1984)
J. Vojtěchovskỳ, K. Chu, J. Berendzen, R.M. Sweet, I. Schlichting, Crystal structures of myoglobin-ligand complexes at near-atomic resolution. Biophys. J. 77(4), 2153–2174 (1999)
Z. Wang, Y. Ando, A.D. Nugraheni, C. Ren, S. Nagao, S. Hirota, Self-oxidation of cytochrome c at methionine80 with molecular oxygen induced by cleavage of the met-heme iron bond. Mol. Biosyst. 10(12), 3130–3137 (2014)
F. Yang, G.N. Phillips Jr., Crystal structures of CO-, deoxy-and met-myoglobins at various pH values. J. Mol. Biol. 256(4), 762–774 (1996)
Acknowledgements
This work was supported in part by Bulgarian Science Fund (Grant DNTS-CN-01/9/2014), Vetenskapsrådet (Sweden), Carl Trygger’s Stiftelse and Qian Ren Grant at Beijing Institute of Technology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hou, Y., Niemi, A.J., Peng, X. et al. Myoglobin ligand gate mechanism analysis by a novel 3D visualization technique. J Math Chem 57, 1586–1597 (2019). https://doi.org/10.1007/s10910-019-01021-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10910-019-01021-4