Abstract
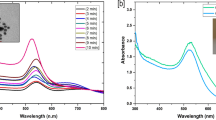
Citrus Tristeza virus (CTV) is one of the most destructive pathogens worldwide that exist as a mixture of malicious (Sever) and tolerable (Mild) strains. Mild strains of CTV can be used to immunize healthy plants from more Severe strains damage. Recently, innovative methods based on the fluorescent properties of DNA/silver nanoclusters have been developed for molecular detection purposes. In this study, a simple procedure was followed to create more active DNA/AgNCs probe for accurate and selective detection of Tristeza Mild-RNA. To this end, four distinct DNA emitter scaffolds (C12, Red, Green, Yellow) were tethered to the Mild capture sequence and investigated in various buffers in order to find highly emissive combinations. Then, to achieve specific and reliable results, several chemical additives, including organic solvents, PEG and organo-soluble salts were used to enhance control fluorescence signals and optimize the hybridization solution. The data showed that, under adjusted conditions, the target sensitivity is enhanced by a factor of five and the high discrimination between Mild and Severe RNAs were obtained. The emission ratio of the DNA/AgNCs was dropped in the presence of target RNAs and I0/I intensity linearly ranged from 1.5 × 10−8 M to 1.8 × 10−6 M with the detection limit of 4.3 × 10−9 M.




Similar content being viewed by others
References
Bar-Joseph M, Batuman O, Roistacher CN (2010) The history to citrus tristeza virus – revisited. In: Complex CTV, Diseases T (eds) Karasev AV, Hilf ME. American Phytopathological Society, St. Paul, MN, pp. 3–26
Garnsey SM, Civerolo EL, Gumpf DJ, Paul C, Hilf ME, Lee RF, Brlansky RH, Yokomi RK (2005) Biological characterization of an international collection of Citrus tristeza virus (CTV) isolates, Proc 16th Congr Int Org Citrus Virologists 75–93.
Lee RF, Niblett CL (2000) Citrus tristeza virus: strains, mild strain cross protection and other management strategies. Rev Hortic Mex 8:25–35
Roistacher CN, da Graça JV, Müller GW (2010) Cross protection against Citrus tristeza virus – a review, in Proceedings of the Seventeenth Conference International Organization of Citrus Virologists, eds Hilf ME, Timmer LW, Milne RG, da Graça JV, (Riverside, CA:IOCV), 3–27
Permar TA, Garnsey SM, Gumpf DJ, Lee RF (1990) A monoclonal-antibody that discriminates strains of citrus tristeza virus. Phytopathology 80:224–228
Narváez G, Skander BS, Ayllón MA, Rubio L, Guerri J, Moreno P (2000) A new procedure to differentiate citrus tristeza virus solates by hybridisation with digoxigenin-labelled cDNA probes. J Virol Methods 85:83–92
Gillings M, Broadbent P, Indsto J, Lee R (1993) Characterization of isolates and strains of citrus tristeza closterovirus using restriction analysis of the coat protein gene amplified by the polymerase chain reaction. J Virol Methods 44:305–317
Ayllón MA, Rubio L, Sentandreu V, Moya A, Guerri J, Moreno P (2006) Variations in two gene sequences of citrus tristeza virus after host passage. Virus Genes 32:119–128
Cheng X, Chen G, Rodriguez WR (2009) Micro-and nanotechnology for viral detection. Anal Bioanal Chem 393:487–501
Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449:851–861
Dadmehr M, Hosseini M, Hosseinkhani S, Ganjali MR, Sheikhnejad R (2015) Label free colorimetric and fluorimetric direct detection of methylated DNA based on silver nanoclusters for cancer early diagnosi. Biosens Bioelectron 73:108–113
Ahmadzadeh Kermani H, Hosseini M, Dadmehr M, Ganjali MR (2016) Rapid restriction enzyme free detection of DNA methyltransfarase activity based on DNA-templated silver nanoclusters. Anal Bioanal Chem 16:4311–4318
Budowle B, Allard MW, Wilson MR, Chakraborty R (2003) Forensics and mitochondrial DNA: applications, debates, and foundations. Annu Rev Genomics Hum Genet 4:119–141
Liu YQ, Zhang M, Yin BC, Ye BC (2012) Attomolar ultrasensitive microRNA detection by DNA-scaffolded silver-nanocluster probe based on isothermal amplification. Anal Chem 84:5165–5169
Yang SW, Vosch T (2011) Rapid detection of microRNA by a silver nanocluster DNA probe. Anal Chem 83:6935–6939
Obliosca JM, Liu C, Batson RA, Babin MC, Werner J (2013) DNA/RNA detection using DNA-templated few-atom silver nanoclusters. Biosensors 3:185–200
Yeh HC, Sharma J, Han JJ, Martinez JS, Werner JH (2011) A beacon of light—a new molecular probe for homogeneous detection of nucleic acid targets. IEEE Nanotechnol Mag 5:28–33
Yeh HC, Sharma J, Yoo H, Martinez JS, Werner JH (2010) Photophysical characterization of fluorescent metal nanoclusters synthesized using oligonucleotides, proteins and small molecule ligands. Proc SPIE 7576:75760 N1–75760 N9
Le Guevel X, Spies C, Daum N, Jung G, Schneider M (2012) Highly fluorescent silver nanoclusters stabilized by glutathione: a promising fluorescent label for bioimaging. Nano Res 5:379–387
Vosch T, Antoku Y, Hsiang JC, Richards CI, Gonzalez JI, Dickson RM (2007) Strongly emissive individual DNA-encapsulated Ag nanoclusters as single-molecule fluorophores. Proc Natl Acad Sci U S A 104:12616–12621
Richards CI, Choi S, Hsiang JC, Antoku Y, Vosch T, Bongiorno A, Tzeng YL, Dickson, RM (2008) Oligonucleotide-stabilized Ag nanocluster fluorophores. J Am Chem Soc 130(15):5038–5039
Sengupta B, Corley C, Cobb K, Saracino A, Jockusch S (2016) DNA scaffolded silver clusters: a critical study. Molecules 21:216
Hames BD, Higgins SJ (1985) Nucleic acid hybridisation: a practical approach. IRL Press, Oxford
Semenov MA, Bolbukh TV, Krasnitskaya AA, Ya Maleyev V (1994) a study of hydration and structural state of human dna during long term storage. Probelems Cryobiol No 4
Ugwu SO, Apte SP (2004) The effect of buffers on protein conformational stability. Pharm Technol 28:86–113
Zhao YQ, Fu JY, Liang AH, Yang BS (2009) The characterization for the binding of calcium and terbium to Euplotes octocarinatus centrin. Spectrochim Acta A 71:1756–1761
Wenner JR, Bloomfield VA (1999) Buffer effects on EcoRV kinetics as measured by fluorescent staining and digital imaging of plasmid cleavage. Anal Biochem 268:201–212
Wolfe AR, Meehan T (1994) The effect of sodium ion concentration on intrastrand base-pairing in single-stranded DNA. Nucleic Acids Res 22(15):3147–3150
Roy KB, Antony T, Sakena A, Bohidar HB (1999) Ethanol-induced condensation of calf thymus DNA studied by laser light scattering. J Phys Chem B 103:5117–5121
Hardwidge P, Pang YP, Zimmerman J, Vaghefi M, Hogrefe R, Maher LJ (2004) Phosphate crowding and DNA bending. In: Mohanty U, Stellwagen N (eds) Curvature and deformation of nucleic acids: recent advances, new paradigms. Am Chem Soc Symposium Series, vol 884. Oxford University Press, New York, NY
Forster S, Schmidt M (1995) Polyelectrolytes in solution. Adv Polym Sci 120:51–133
Agbavwe C, Somoza MM (2011) Sequence-dependent fluorescence of cyanine dyes on microarrays. PLoS One 6:e22177
Ke FY, Luu YK, Liang DH (2010) Characterizing DNA condensation and conformational changes in organic solvents. PLoS One 5:e13308
Kypr J, Kejnovska I, Renciuk D, Vorlíckova M (2009) Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res 37:1713–1725
Ellis RJ (2001) Macromolecular crowding: obvious, but underappreciated. Trends Biochem Sci 26:597–604
Hou S, Trochimczyk P, Sun L, Wisniewska A, Kalwarczyk T, Zhang X, Wielgus-Kutrowska B, Bzowska A, Holyst R (2016) How can macromolecular crowding inhibit biological reactions? The enhanced formation of DNA nanoparticles. Sci Rep 6
Knowles DB, LaCroix AS, Deines NF, Shkel I, Record MT (2011) Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. J Proc Natl Acad Sci USA 108:12699–12704
Luo C, Zhang Y, Zeng X, Zeng Y, Wang Y (2005) The role of poly(ethylene glycol) in the formation of silver nanoparticles. J Colloid Interface Sci 288:444–448
Kaneta T, Tanaka S, Yoshida H (1991) Improvement of resolution in the capillary electrophoretic separation of catecholamines by complex formation with boric acid and control of electroosmosis with a cationic surfactant. J Chromatogr A 538:385–391
Buchmueller KL, Weeks KM (2004) Tris–borate is a poor counterion for RNA: a cautionary tale for RNA folding studies. Nucleic Acids Res 32:e184
Williams LD, Maher LJ (2000) Electrostatic mechanisms of DNA deformation. Annu Rev Biophys Biomol Struct 29:497–521
Ruiz-Ruiz S, Moreno P, Guerri J, Ambrós S (2007) A real-time RT-PCR assay for detection and absolute quantification of Citrus tristeza virus in different plant tissues. J Virol Methods 145:96–105
Acknowledgments
The authors would like to acknowledge the financial support of Science and Technology Park of University of Tehran for this research under grant number of (94056).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shokri, E., Hosseini, M., Faridbod, F. et al. Synthesis and Assessment of DNA/Silver Nanoclusters Probes for Optimal and Selective Detection of Tristeza Virus Mild Strains. J Fluoresc 26, 1795–1803 (2016). https://doi.org/10.1007/s10895-016-1871-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-016-1871-7