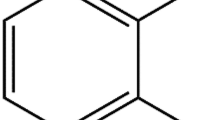
Abstract
A novel method for the quantitative assessment of the membrane partitioning of a ligand from the aqueous phase is described, demonstrated here with the thoroughly studied antipsychotic chlorpromazine (CPZ). More specifically, collisional quenching of the fluorescence of a pyrene labeled fluorescent lipid analog 1-palmitoyl-2[10-(pyren-1-yl)]decanoyl-sn-glycero-3-phosphocholine (PPDPC) by CPZ was utilized, using 1-palmitoyl-2-oleyl-sn-glycero-3-phosphocholine and -serine (POPC and POPS) liposomes as model membranes. The molar partition coefficient is obtained from two series of titrations, one with constant [phospholipid] and increasing [drug] and the other with constant [drug] and varying total [phospholipid], the latter further comprising of large unilamellar vesicles (LUVs) of POPC/POPS/PPDPC at a constant concentration of 10 μM and indicated concentrations of POPC/POPS LUVs. Notably, the approach described is generic and can be employed in screening for the membrane partitioning of compounds, providing that a suitable fluorescence parameter can be incorporated into one population of liposomes utilized as model membranes.




Similar content being viewed by others
References
Kansy M, Senner F, Gubernator K (1998) Physicochemical high throughput screening: Parallel artificial membrane permeation assay in the description of passive absorption processes. J Med Chem 41(7):1007–1010
Schreier S, Malheiros SV, de Paula E (2000) Surface active drugs: Self-association and interaction with membranes and surfactants. physicochemical and biological aspects. Biochim Biophys Acta 1508(1–2):210–234
Santos NC, Prieto M, Castanho MARB (2003) Quantifying molecular partition into model systems of biomembranes: An emphasis on optical spectroscopic methods. Biochim Biophys Acta, Biomem 1612(2):123–135
Suomalainen P, Johans C, Söderlund T, Kinnunen PK (2004) Surface activity profiling of drugs applied to the prediction of blood-brain barrier permeability. J Med Chem 47(7):1783–1788
Mycek MJ (1997) Pharmacology 2nd ed. Lippincott-Raven, Philadelphia, PA
Seeman P (1972) The membrane actions of anesthetics and tranquilizers. Pharmacol Rev 24(4):583–655
Zhao H, Mattila JP, Holopainen JM, Kinnunen PK (2001) Comparison of the membrane association of two antimicrobial peptides, magainin 2 and indolicidin. Biophys J 81(5):2979–2991
Jutila A, Rytömaa M, Kinnunen PK (1998) Detachment of cytochrome c by cationic drugs from membranes containing acidic phospholipids: Comparison of lidocaine, propranolol, and gentamycin. Mol Pharmacol 54(4):722–732
Söderlund T, Lehtonen JY, Kinnunen PK (1999) Interactions of cyclosporin A with phospholipid membranes: Effect of cholesterol. Mol Pharmacol 55(1):32–38
Antkowiak B (2001) How do general anaesthetics work? Naturwissenschaften 88(5):201–213
Conrad MJ, Singer SJ (1981) The solubility of amphipathic molecules in biological membranes and lipid bilayers and its implications for membrane structure. Biochemistry 20(4):808–818
Luxnat M, Muller HJ, Galla HJ (1984) Membrane solubility of chlorpromazine. hygroscopic desorption and centrifugation methods yield comparable results. Biochem J 224(3):1023–1026
Jutila A, Söderlund T, Pakkanen AL, Huttunen M, Kinnunen PK (2001) Comparison of the effects of clozapine, chlorpromazine, and haloperidol on membrane lateral heterogeneity. Chem Phys Lipids 112(2):151–163
Bretscher (1972) Asymmetrical lipid bilayer structure for biological membranes. Nature: New biology 236(61):11
Dolis D, Moreau C, Zachowski A, Devaux PF (1997) Aminophospholipid translocase and proteins involved in transmembrane phospholipid traffic. Biophys Chem 68(1–3):221–231
Devaux PF (1991) Static and dynamic lipid asymmetry in cell membranes. Biochemistry 30(5):1163–1173
Nerdal W, Gundersen SA, Thorsen V, Hoiland H, Holmsen H (2000) Chlorpromazine interaction with glycerophospholipid liposomes studied by magic angle spinning solid state (13)C-NMR and differential scanning calorimetry. Biochim Biophys Acta 1464(1):165–175
Underhaug Gjerde A, Holmsen H, Nerdal W (2004) Chlorpromazine interaction with phosphatidylserines: A (13)C and (31)P solid-state NMR study. Biochim Biophys Acta 1682(1–3):28–37
Varnier Agasoster A, Holmsen H (2001) Chlorpromazine associates with phosphatidylserines to cause an increase in the lipid’s own interfacial molecular area–role of the fatty acyl composition. Biophys Chem 91(1):37–47
Luxnat M, Galla HJ (1986) Partition of chlorpromazine into lipid bilayer membranes: The effect of membrane structure and composition. Biochim Biophys Acta 856(2):274–282
Takegami S, Kitamura K, Kitade T, et al (2005) Effects of phosphatidylserine and phosphatidylethanolamine content on partitioning of triflupromazine and chlorpromazine between phosphatidylcholine-aminophospholipid bilayer vesicles and water studied by second-derivative spectrophotometry. Chem Pharm Bull (Tokyo) 53(1):147–150
Tehrani S, Brandstater N, Saito YD, Dea P (2001) Studies on the size and stability of chlorpromazine hydrochloride nanostructures in aqueous solution. Biophys Chem 94(1–2):87–96
Chen JY, Brunauer LS, Chu FC, Helsel CM, Gedde MM, Huestis WH (2003) Selective amphipathic nature of chlorpromazine binding to plasma membrane bilayers. Biochim Biophys Acta 1616(1):95–105
MacDonald RC, MacDonald RI, Menco BP, Takeshita K, Subbarao NK, Hu LR (1991) Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. Biochim Biophys Acta 1061(2):297–303
Kucerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF (2005) Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys J 88(4):2626–2637
Metso AJ, Jutila A, Mattila J, Holopainen JM, Kinnunen PKJ (2003) Nature of the main transition of dipalmitoylphosphocholine bilayers inferred from fluorescence spectroscopy. J Phys Chem B 107(5):1251–1257
Lakowicz JR (1999) Principles of Fluorescence Spectroscopy 2nd ed. Kluwer Academic/Plenum, New York
Rytömaa M, Kinnunen PK (1994) Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. J Biol Chem 269(3):1770–1774
Mustonen P, Kinnunen PK (1993) On the reversal by deoxyribonucleic acid of the binding of adriamycin to cardiolipin-containing liposomes. J Biol Chem 268(2):1074–1080
Koiv A, Palvimo J, Kinnunen PK (1995) Evidence for ternary complex formation by histone H1, DNA, and liposomes. Biochemistry 34(25):8018–8027
Acknowledgments
The authors wish to thank DDS Antti Pakkanen for estimating the molar volume of CPZ. The technical assistance of Kristiina Söderholm and Kaija Niva is highly appreciated. HBBG is supported by the Finnish Academy and Sigrid Juselius Foundation. JMA is supported by the Research Foundation of Orion Corporation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Parry, M.J., Jutila, A., Kinnunen, P.K.J. et al. A Versatile Method for Determining the Molar Ligand-Membrane Partition Coefficient. J Fluoresc 17, 97–103 (2007). https://doi.org/10.1007/s10895-006-0138-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-006-0138-0