Abstract
MALT1 (mucosa-associated lymphoid tissue lymphoma-translocation gene 1) is an intracellular signaling protein that activates NFκB and is crucial for both the adaptive and innate immune responses. Only 6 patients with immune deficiencies secondary to inherited mutations in the MALT1 gene have been described.
Purpose
To provide clinical and immunological insights from 2 patients diagnosed with MALT1 immunodeficiency syndrome due to a novel MALT1 mutation.
Methods
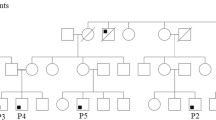
Two cousins with suspected combined immunodeficiency underwent immunological and genetic work-up, including lymphocyte phenotyping, lymphocyte activation by mitogen stimulation, and next-generation sequencing (NGS) of T cell receptor gamma chain (TRG) repertoire. Whole exome sequencing was performed to identify the underlying genetic defect.
Results
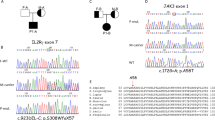
Clinical findings included recurrent infections, failure to thrive, lymphadenopathy, dermatitis, and autoimmunity. Immune work-up revealed lymphocytosis, low to normal levels of immunoglobulins, absence of regulatory T cells, and low Th17 cells. A normal proliferative response was induced by phytohemagglutinin and IL-2 but was diminished with anti-CD3. TRG repertoire was diverse with a clonal expansion pattern. Genetic analysis identified a novel autosomal recessive homozygous c.1799T>A; p. I600N missense mutation in MALT1. MALT1 protein expression was markedly reduced, and in vitro IL-2 production and NFκB signaling pathway were significantly impaired.
Conclusions
Two patients harboring a novel MALT1 mutation presented with signs of immune deficiency and dysregulation and were found to have an abnormal T cell receptor repertoire. These findings reinforce the link between MALT1 deficiency and combined immunodeficiency. Early diagnosis is crucial, and curative treatment by hematopoietic stem cell transplantation may be warranted.








Similar content being viewed by others
Abbreviations
- CBM:
-
CARD9/10/11-BCL10-MALT1
- CID:
-
combined immunodeficiency
- HSCT:
-
hematopoietic stem cell transplantation
- MALT-1:
-
mucosa-associated lymphoid tissue lymphoma-translocation gene 1
- NGS:
-
next-generation sequencing
- PBMCs:
-
peripheral blood mononuclear cells
- SCID:
-
severe combined immunodeficiency
- TRECs:
-
T cell receptor excision circles
- TCR:
-
T cell receptor
References
Tasher D, Dalal I. The genetic basis of severe combined immunodeficiency and its variants. Appl Clin Genet. 2012;5:67–80.
Somech R, Roifman CM. Mutation analysis should be performed to rule out gammac deficiency in children with functional severe combined immune deficiency despite apparently normal immunologic tests. J Pediatr. 2005;147(4):555–7.
Picard C, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, et al. Primary immunodeficiency diseases: an update on the Classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J Clin Immunol. 2015;35(8):696–726.
Roifman CM, Somech R, Kavadas F, Pires L, Nahum A, Dalal I, et al. Defining combined immunodeficiency. J Allergy Clin Immunol. 2012;130(1):177–83.
Speckmann C, Doerken S, Aiuti A, Albert MH, Al-Herz W, Allende LM, et al. A prospective study on the natural history of patients with profound combined immunodeficiency: an interim analysis. J Allergy Clin Immunol. 2017;139(4):1302–10 e4.
Akar HH, Patiroglu T, Hershfield M, van der Burg M. Combined immunodeficiencies: twenty years experience from a single center in Turkey. Cent Eur J Immunol. 2016;41(1):107–15.
Jabara HH, Ohsumi T, Chou J, Massaad MJ, Benson H, Megarbane A, et al. A homozygous mucosa-associated lymphoid tissue 1 (MALT1) mutation in a family with combined immunodeficiency. J Allergy Clin Immunol. 2013;132(1):151–8.
McKinnon ML, Rozmus J, Fung SY, Hirschfeld AF, Del Bel KL, Thomas L, et al. Combined immunodeficiency associated with homozygous MALT1 mutations. J Allergy Clin Immunol. 2014;133(5):1458–62 62 e1–7.
Punwani D, Wang H, Chan AY, Cowan MJ, Mallott J, Sunderam U, et al. Combined immunodeficiency due to MALT1 mutations, treated by hematopoietic cell transplantation. J Clin Immunol. 2015;35(2):135–46.
Charbit-Henrion F, Jeverica AK, Begue B, Markelj G, Parlato M, Avcin SL, et al. Deficiency in mucosa-associated lymphoid tissue lymphoma translocation 1: a novel cause of IPEX-like syndrome. J Pediatr Gastroenterol Nutr. 2017;64(3):378–84.
Ruland J, Duncan GS, Wakeham A, Mak TW. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 2003;19(5):749–58.
Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4(5):348–59.
Thome M. Multifunctional roles for MALT1 in T-cell activation. Nat Rev Immunol. 2008;8(7):495–500.
Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2(9):a003004.
Lu HY, Bauman BM, Arjunaraja S, Dorjbal B, Milner JD, Snow AL, et al. The CBM-opathies—a rapidly expanding spectrum of human inborn errors of immunity caused by mutations in the CARD11-BCL10-MALT1 complex. Front Immunol. 2018;9:2078.
Juilland M, Thome M. Holding all the CARDs: how MALT1 controls CARMA/CARD-dependent signaling. Front Immunol. 2018;9:1927.
Hamilton KS, Phong B, Corey C, Cheng J, Gorentla B, Zhong X, et al. T cell receptor-dependent activation of mTOR signaling in T cells is mediated by Carma1 and MALT1, but not Bcl10. Sci Signal. 2014;7(329):ra55.
E A, P D, MP L, V G. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604.
CJ K. Simpson diversity and the Shannon–Wiener index as special cases of a generalized entropy. Oikos. 2005;109:203–7.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
Li MX, Gui HS, Kwan JS, Bao SY, Sham PC. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 2012;40(7):e53.
Ma CA, Stinson JR, Zhang Y, Abbott JK, Weinreich MA, Hauk PJ, et al. Germline hypomorphic CARD11 mutations in severe atopic disease. Nat Genet. 2017;49(8):1192–201.
Sufficool KE, Lockwood CM, Abel HJ, Hagemann IS, Schumacher JA, Kelley TW, et al. T-cell clonality assessment by next-generation sequencing improves detection sensitivity in mycosis fungoides. J Am Acad Dermatol. 2015;73(2):228–36 e2.
Kansal R, Grody WW, Zhou J, Dong L, Li X. The value of T-cell receptor gamma (TRG) clonality evaluation by next-generation sequencing in clinical hematolymphoid tissues: a descriptive study of 41 cases from a single institution. Am J Clin Pathol. 2018;150:193–223.
Mahe E, Pugh T, Kamel-Reid S. T cell clonality assessment: past, present and future. J Clin Pathol. 2018;71(3):195–200.
Cruz MS, Diamond A, Russell A, Jameson JM. Human alphabeta and gammadelta T cells in skin immunity and disease. Front Immunol. 2018;9:1304.
Johnston EL, Roberts DA. Contaminants reduce the richness and evenness of marine communities: a review and meta-analysis. Environ Pollut. 2009;157(6):1745–52.
Simpson EH. Measurement of diversity. Nature. 1949;163:688.
Rechavi E, Lev A, Lee YN, Simon AJ, Yinon Y, Lipitz S, et al. Timely and spatially regulated maturation of B and T cell repertoire during human fetal development. Sci Transl Med. 2015;7(276):276ra25.
Lee YN, Frugoni F, Dobbs K, Tirosh I, Du L, Ververs FA, et al. Characterization of T and B cell repertoire diversity in patients with RAG deficiency. Sci Immunol. 2016;1(6):eaah6109.
Bornancin F, Renner F, Touil R, Sic H, Kolb Y, Touil-Allaoui I, et al. Deficiency of MALT1 paracaspase activity results in unbalanced regulatory and effector T and B cell responses leading to multiorgan inflammation. J Immunol. 2015;194(8):3723–34.
Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302(5650):1581–4.
Isaacson P, Wright DH. Malignant lymphoma of mucosa-associated lymphoid tissue. A distinctive type of B-cell lymphoma. Cancer. 1983;52(8):1410–6.
Willis TG, Jadayel DM, Du MQ, Peng H, Perry AR, Abdul-Rauf M, et al. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell. 1999;96(1):35–45.
Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno-Garcia ME, Ovechkina YL, et al. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 2005;23(6):561–74.
Lucas PC, Yonezumi M, Inohara N, McAllister-Lucas LM, Abazeed ME, Chen FF, et al. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J Biol Chem. 2001;276(22):19012–9.
Juilland M, Thome M. Role of the CARMA1/BCL10/MALT1 complex in lymphoid malignancies. Curr Opin Hematol. 2016;23(4):402–9.
Perez de Diego R. Immunodeficiency and CARD-BCL10-MALT1. Oncotarget. 2015;6(24):19934–5.
Dadi H, Jones TA, Merico D, Sharfe N, Ovadia A, Schejter Y, et al. Combined immunodeficiency and atopy caused by a dominant negative mutation in caspase activation and recruitment domain family member 11 (CARD11). J Allergy Clin Immunol. 2018;141(5):1818–30 e2.
Dorjbal B, Stinson JR, Ma CA, Weinreich MA, Miraghazadeh B, Hartberger JM, et al. Hypomorphic caspase activation and recruitment domain 11 (CARD11) mutations associated with diverse immunologic phenotypes with or without atopic disease. J Allergy Clin Immunol. 2018.
Yu JW, Hoffman S, Beal AM, Dykon A, Ringenberg MA, Hughes AC, et al. MALT1 protease activity is required for innate and adaptive immune responses. PLoS One. 2015;10(5):e0127083.
Gewies A, Gorka O, Bergmann H, Pechloff K, Petermann F, Jeltsch KM, et al. Uncoupling Malt1 threshold function from paracaspase activity results in destructive autoimmune inflammation. Cell Rep. 2014;9(4):1292–305.
Shearer WT, Rosenblatt HM, Gelman RS, Oyomopito R, Plaeger S, Stiehm ER, et al. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J Allergy Clin Immunol. 2003;112(5):973–80.
Schatorje EJ, Gemen EF, Driessen GJ, Leuvenink J, van Hout RW, de Vries E. Paediatric reference values for the peripheral T cell compartment. Scand J Immunol. 2012;75(4):436–44.
Piatosa B, Wolska-Kusnierz B, Pac M, Siewiera K, Galkowska E, Bernatowska E. B cell subsets in healthy children: reference values for evaluation of B cell maturation process in peripheral blood. Cytometry B Clin Cytom. 2010;78(6):372–81.
Acknowledgements
The authors thank the patients and their families for their collaboration and the expert care by the interdisciplinary pediatric teams. Raz Somech is supported by grants from the Jeffrey Modell Foundation (JMF) and the Israeli Science Foundation (ISF). The authors would like to acknowledge Dr. Abigail Lazar for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declared that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Frizinsky, S., Rechavi, E., Barel, O. et al. Novel MALT1 Mutation Linked to Immunodeficiency, Immune Dysregulation, and an Abnormal T Cell Receptor Repertoire. J Clin Immunol 39, 401–413 (2019). https://doi.org/10.1007/s10875-019-00629-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-019-00629-0