Abstract
Betaine aldehyde dehydrogenase (BADH) catalyzes the oxidation of betaine aldehyde to glycine betaine using NAD+ as a coenzyme. Studies in porcine kidney BADH (pkBADH) suggested that the enzyme exhibits heterogeneity of active sites and undergoes potassium-induced conformational changes. This study aimed to analyze if potassium concentration plays a role in the heterogeneity of pkBADH active sites through changes in NAD+ affinity constants, in its secondary structure content and stability. The enzyme was titrated with NAD+ 1 mM at fixed-variable KCl concentration, and the interaction measured by Isothermal Titration Calorimetry (ITC) and Circular Dichroism (CD). ITC data showed that K+ increased the first active site affinity in a manner dependent on its concentration; KD values to the first site were 14.4, 13.1, and 10.4 μM, at 25, 50, and 75 mM KCl. ΔG values showed that the coenzyme binding is a spontaneous reaction without changes between active sites or depending on KCl concentration. ΔH and TΔSb values showed that NAD+ binding to the active site is an endothermic process and is carried out at the expense of changes in entropy. α-Helix content increased as KCl increased, enzyme (Tm)app values were 2.6 °C and 3.3 °C higher at 20 mM and 200 mM K+. PkBADH molecular model showed three different interaction K+ sites. Results suggested K+ can interact with pkBADH and cause changes in the secondary structure, it provokes changes in the enzyme affinity by the coenzyme, and in the thermostability.





Similar content being viewed by others
References
Ahvazi B, Coulombe R, Delarge M, Vedadi M, Zhang L, Meighen E, Vrielink A (2000) Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity. Biochem J 349(3):853–861
Ayala-Castro HG, Valenzuela-Soto EM, Figueroa-Soto CG, Munoz-Clares RA (2007) Complex, unusual conformational changes in kidney betaine aldehyde dehydrogenase suggested by chemical modification with disulfiram. Arch Biochem Biophys 468(2):167–173
Bezerra GA, Ohara-Nemoto Y, Cornaciu I, Fedosyuk S, Hoffmann G, Round A, Márquez JA, Nemoto TK, Djinović-Carugo K (2017) Bacterial protease uses distinct thermodynamic signatures for substrate recognition. Sci Rep 7(1):2848
Black S (1951) Yeast aldehyde dehydrogenase. Arch Biochem Biophys 34(1):86–97
Cobessi D, Tête-Favier F, Marchal S, Branlant G, Aubry A (2000) Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J Mol Biol 300(1):141–152
Delgado-Gaytán MF, Rosas-Rodríguez JA, Yepiz-Plascencia G, Figueroa-Soto CG, Valenzuela-Soto EM (2017) Cloning and molecular characterization of the betaine aldehyde dehydrogenase involved in the biosynthesis of glycine betaine in white shrimp (Litopenaeus vannamei). Chem Biol Interact 276:65–74
Ferrante A, Gorski J (2012) Enthalpy–entropy compensation and cooperativity as thermodynamic epiphenomena of structural flexibility in ligand–receptor interactions. J Mol Biol 417(5):454–467
Figueroa-Soto CG, Valenzuela-Soto EM (2000) Kinetic study of porcine kidney betaine aldehyde dehydrogenase. Biochem Biophys Res Commun 269:596–603
Fox JM, Zhao M, Fink MJ, Kang K, Whitesides GM (2018) The molecular origin of enthalpy/entropy compensation in biomolecular recognition. Annu Rev Biophys 47:223–250
Garza-Ramos G, Carrillo-Nava E, Costas M, Mújica-Jiménez C (2007) Thermal stability of Betaine aldehyde dehydrogenase. In: Weiner H, Plapp B, Lindhal R, Maser E (eds) Enzymology and molecular biology of carbonyl metabolism 13. Purdue University Press, West Lafayette, pp 64–76
Garza-Ramos G, Mújica-Jiménez C, Muñoz-Clares RA (2013) Potassium and ionic strength effects on the conformational and thermal stability of two aldehyde dehydrogenases reveal structural and functional roles of K+-binding sites. PLoS One 8(1):e54899
González-Segura L, Velasco-García R, Muñoz-Clares RA (2002) Modulation of the reactivity of the essential cysteine residue of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa. Biochem J 361(3):577–585
González-Segura L, Rudiño-Piñera E, Muñoz-Clares RA, Horjales E (2009) The crystal structure of a ternary complex of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa provides new insight into the reaction mechanism and shows a novel binding mode of the 2′-phosphate of NADP+ and a novel cation binding site. J Mol Biol 385(2):542–557
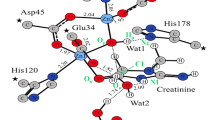
González-Segura L, Riveros-Rosas H, Díaz-Sánchez ÁG, Julián-Sánchez A, Muñoz-Clares RA (2013) Potential monovalent cation-binding sites in aldehyde dehydrogenases. Chem Biol Interact 202(1):41–50
Halavaty AS, Rich RL, Chen C, Joo JC, Minasov G, Dubrovska I, et al (2015) Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus. Acta Crystallogr D Biol Crystallogr 71(5):1159–1175
Hurley TD, Perez-Miller S, Breen H (2001) Order and disorder in mitochondrial aldehyde dehydrogenase. Chem Biol Interact 130:3–14
Johansson K, Ramaswamy S, Eklund H, El-Ahmad M, Hjelmqvist L, Jörnvall H (1998) Structure of betaine aldehyde dehydrogenase at 2.1 Å resolution. Protein Sci 7(10):2106–2117
Končitíková R, Vigouroux A, Kopečná M, Šebela M, Moréra S, Kopečný (2019) Kinetic and structural analysis of human ALDH9A1. Biosci Rep:BSR20190558
Lamb AL, Newcomer ME (1999) The structure of retinal dehydrogenase type II at 2.7 Å resolution: implications for retinal specificity. Biochemistry 38(19):6003–6011
Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, Kuo I, Perozich J, Lindahl R, Hempel J (1997) The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD+ and the Rossmann fold. Nat Struct Mol Biol 4(4):317–326
Molnes J, Teigen K, Aukrust I, Bjørkhaug L, Søvik O, Flatmark T, Njølstad PR (2011) Binding of ATP at the active site of human pancreatic glucokinase–nucleotide-induced conformational changes with possible implications for its kinetic cooperativity. FEBS J 278(13):2372–2386
Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN (1998) Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure 6(12):1541–1551
Muñoz-Bacasehua C, Rosas-Rodríguez JA, Arvizu-Flores AA, Stephens-Camacho A, Soñanez-Organis JG, Figueroa-Soto CG and Valenzuela-Soto EM (2019) Heterogeneity of active sites in porcine kidney betaine aldehyde dehydrogenase is modulated by potassium. J Mol Recognit (Submitted)
Muñoz-Clares RA, González-Segura L, Mújica-Jiménez C, Contreras-Díaz L (2003) Ligand-induced conformational changes of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa and Amaranthus hypochondriacus L. leaves affecting the reactivity of the catalytic thiol. Chem Biol Interact 143:129–137
Steinmetz CG, Xie P, Weiner H, Hurley TD (1997) Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure 5(5):701–711
Stines-Chaumeil C, Talfournier F, Branlant G (2006) Mechanistic characterization of the MSDH (methylmalonate semialdehyde dehydrogenase) from Bacillus subtilis. Biochem J 395(1):107–115
Testore G, Cravanzola C, Bedino S (1999) Aldehyde dehydrogenase from rat intestinal mucosa: purification and characterization of an isozyme with high affinity for γ-aminobutyraldehyde. Int J Biochem Cell Biol 31(7):777–786
Valenzuela-Soto EM, Velasco-García R, Mújica-Jiménez C, Muñoz-Clares RA (2003) Monovalent cations requirements for the stability of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa, porcine kidney and amaranth leaves. Chem Biol Interact 143:139–148
Valenzuela-Soto EM, Ayala-Castro H, Muñoz-Clares R, Maser E, Plapp B, Lindahl R, Weiner H (2005) Effects of monovalent and divalent cations on the thermostability of porcine kidney betaine aldehyde dehydrogenase. In: Weiner H, Plapp B, Lindhal R, Maser E (eds) Enzymology and molecular biology of carbonyl metabolism 12. Purdue University Press, West Lafayette, pp 104–109
Vasiliou V, Pappa A, Petersen DR (2000) Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem Biol Interact 129(1):1–19
Velasco-García R, Mújica-Jiménez C, Mendoza-Hernández G, Muñoz-Clares RA (1999) Rapid purification and properties of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa. J Bacteriol 181(4):1292–1300
Velasco-García R, Gonzáles-Segura L y Muñoz-Clares RA (2000) Steady-state kinetic mechanism of the NADP+-and NAD+-dependent reactions catalysed by betaine aldehyde dehydrogenase from Pseudomonas aeruginosa. Biochem J 352(3):675–683
Von Tigerstrom RG, Razzell W (1968) Aldehyde dehydrogenase I. purification and properties of the enzyme from Pseudomonas aeruginosa. J Biol Chem 243(10):2691–2702
Wang X, Weiner H (1995) Involvement of glutamate 268 in the active site of human liver mitochondrial (class 2) aldehyde dehydrogenase as probed by site-directed mutagenesis. Biochemistry 34(1):237–243
Wang Y, Zhu M, Liu F, Wu X, Pan D, Liu J, Fan S, Wang Z, Tang J, Na R (2016) Comparative studies of interactions between fluorodihydroquinazolin derivatives and human serum albumin with fluorescence spectroscopy. Molecules 21(10):1373
Wei B, Weiner H (2001) Making an oriental equivalent of the yeast cytosolic aldehyde dehydrogenase as well as making one with positive cooperativity in coenzyme binding by mutations of glutamate 492 and arginine 480. Chem Biol Interact 130:173–179
Wei B, Ni L, Hurley TD, Weiner H (2000) Cooperativity in nicotinamide adenine dinucleotide binding induced by mutations of arginine 475 located at the subunit interface in the human liver mitochondrial class 2 aldehyde dehydrogenase. Biochemistry 39(18):5295–5302
Weiner H, Hu J, Sanny CG (1976) Rate-limiting steps for the esterase and dehydrogenase reaction catalyzed by horse liver aldehyde dehydrogenase. J Biol Chem 251(13):3853–3855
Acknowledgments
César Muñoz-Bacasehua gratefully acknowledges a scholarship from CONACyT for graduate studies. Authors are grateful to Dr. G. Garza-Ramos for providing facilities and constant support to carry out CD spectra.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Muñoz-Bacasehua, C., Rosas-Rodríguez, J.A., Arvizu-Flores, A.A. et al. Role of potassium levels in pkBADH heterogeneity of NAD+ binding site. J Bioenerg Biomembr 52, 61–70 (2020). https://doi.org/10.1007/s10863-020-09827-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10863-020-09827-7