Abstract
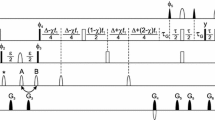
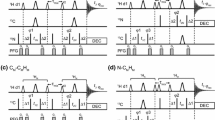
Spectral editing is crucial to simplify the crowded solid-state NMR spectra of proteins. New techniques are introduced to edit 13C-13C correlations of uniformly labeled proteins under moderate magic-angle spinning (MAS), based on our recent frequency-selective homonuclear recoupling sequences [Zhang et al., J. Phys. Chem. Lett. 2020, 11, 8077–8083]. The signals of alanine, serine, or threonine residues are selected out by selective 13Cα-13Cβ double-quantum filtering (DQF). The 13Cα-13Cβ correlations of alanine residues are selectively established with efficiency up to ~ 1.8 times that by dipolar-assisted rotational resonance (DARR). The techniques are shown in 2D/3D NCCX experiments and applied to the uniformly 13C, 15N labeled Aquaporin Z (AqpZ) membrane protein, demonstrating their potential to simplify spectral analyses in biological solid-state NMR.







Similar content being viewed by others
References
Bak M, Rasmussen JT, Nielsen NC (2000) SIMPSON: A general simulation program for solid-state NMR spectroscopy. J Magn Reson 147:296–330
Baldus M, Petkova AT, Herzfeld J, Griffin RG (1998) Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol Phys 95:1197–1207
Castellani F, van Rossum BJ, Diehl A, Rehbein K, Oschkinat H (2003) Determination of solid-state NMR structures of proteins by means of three-dimensional N-15-C-13-C-13 dipolar correlation spectroscopy and chemical shift analysis. Biochemistry 42:11476–11483
Chevelkov V, Giller K, Becker S, Lange A (2013a) Efficient CO-CA transfer in highly deuterated proteins by band-selective homonuclear cross-polarization. J Magn Reson 230:205–211
Chevelkov V, Shi C, Fasshuber HK, Becker S, Lange A (2013b) Efficient band-selective homonuclear CO-CA cross-polarization in protonated proteins. J Biomol NMR 56:303–311
De Paepe G, Lewandowski JR, Griffin RG (2008) Spin dynamics in the modulation frame: application to homonuclear recoupling in magic angle spinning solid-state NMR. J Chem Phys 128:124503
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) Nmrpipe - a multidimensional spectral processing system based on unix pipes. J Biomol NMR 6:277–293
Duong NT, Raran-Kurussi S, Nishiyama Y, Agarwal V (2018) Quantitative H-1-H-1 distances in protonated solids by frequency-selective recoupling at fast magic angle spinning NMR. J Phys Chem Lett 9:5948–5954
Fasshuber HK, Demers JP, Chevelkov V, Giller K, Becker S, Lange A (2015) Specific C-13 labeling of leucine, valine and isoleucine methyl groups for unambiguous detection of long-range restraints in protein solid-state NMR studies. J Magn Reson 252:10–19
Feng X et al (1997) Direct determination of a molecular torsional angle in the membrane protein rhodopsin by solid-state NMR. J Am Chem Soc 119:6853–6857
Franks WT, Kloepper KD, Wylie BJ, Rienstra CM (2007) Four-dimensional heteronuclear correlation experiments for chemical shift assignment of solid proteins. J Biomol NMR 39:107–131
Franks WT et al (2005) Magic-angle spinning solid-state NMR spectroscopy of the beta 1 immunoglobulin binding domain of protein G (GB1): N-15 and C-13 chemical shift assignments and conformational analysis. J Am Chem Soc 127:12291–12305
Fung BM, Khitrin AK, Ermolaev K (2000) An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson 142:97–101
Goobes G, Boender GJ, Vega S (2000) Spinning-frequency-dependent narrowband RF-driven dipolar recoupling. J Magn Reson 146:204–219
Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE (1999) A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated N-15-, C-13-, H-2-labeled proteins. J Biomol NMR 13:369–374
Gullion T, Schaefer J (1989) Rotational-echo double-resonance NMR. J Magn Reson 81:196–200
Hing AW, Vega S, Schaefer J (1992) Transferred-echo double-resonance NMR. J Magn Reson 96:205–209
Hohwy M, Jakobsen HJ, Eden M, Levitt MH, Nielsen NC (1998) Broadband dipolar recoupling in the nuclear magnetic resonance of rotating solids: a compensated C7 pulse sequence. J Chem Phys 108:2686–2694
Hohwy M, Rienstra CM, Griffin RG (2002) Band-selective homonuclear dipolar recoupling in rotating solids. J Chem Phys 117:4973–4987
Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG (1999) Fivefold symmetric homonuclear dipolar recoupling in rotating solids: Application to double quantum spectroscopy. J Chem Phys 110:7983–7992
Hu KN, Tycko R (2009) Zero-quantum frequency-selective recoupling of homonuclear dipole-dipole interactions in solid state nuclear magnetic resonance. J Chem Phys 131:045101
Jehle S, Hiller M, Rehbein K, Diehl A, Oschkinat H, van Rossum BJ (2006a) Spectral editing: selection of methyl groups in multidimensional solid-state magic-angle spinning NMR. J Biomol NMR 36:169–177
Jehle S, Rehbein K, Diehl A, van Rossum BJ (2006b) Amino-acid selective experiments on uniformly C-13 and N-15 labeled proteins by MAS NMR: Filtering of lysines and arginines. J Magn Reson 183:324–328
Kobayashi T, Wang ZR, Pruski M (2019) Homonuclear dipolar recoupling of arbitrary pairs in multi-spin systems under magic angle spinning: a double-frequency-selective ZQ-SEASHORE experiment. Solid State Nucl Magn Reson 101:76–81
Li JP, Her AS, Traaseth NJ (2020) Site-specific resolution of anionic residues in proteins using solid-state NMR spectroscopy. J Biomol NMR 74:355–363
Mao JD, Schmidt-Rohr K (2005) Methylene spectral editing in solid-state C-13 NMR by three-spin coherence selection. J Magn Reson 176:1–6
Nielsen AB, Jain SK, Nielsen NC (2011) Low-power homonuclear dipolar recoupling using supercycled symmetry-based and exponentially-modulated pulse sequences. Chem Phys Lett 503:310–315
Nielsen NC, Bildsoe H, Jakobsen HJ, Levitt MH (1994) Double-quantum homonuclear rotary resonance: efficient dipolar recovery in magic-angle spinning nuclear magnetic resonance. J Chem Phys 101:1805–1812
Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H (2001) Backbone and side-chain C-13 and N-15 signal assignments of the alpha-spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 tesla. ChemBioChem 2:272–281
Qiang W, Tycko R (2012) Zero-quantum stochastic dipolar recoupling in solid state nuclear magnetic resonance. J Chem Phys 137:104201
Raleigh DP, Levitt MH, Griffin RG (1988) Rotational resonance in solid-state NMR. Chem Phys Lett 146:71–76
Schmidt-Rohr K, Fritzsching KJ, Liao SY, Hong M (2012) Spectral editing of two-dimensional magic-angle-spinning solid-state NMR spectra for protein resonance assignment and structure determination. J Biomol NMR 54:343–353
Schmidt-Rohr K, Mao JD (2002) Efficient CH-group selection and identification in C-13 solid-state NMR by dipolar DEPT and H-1 chemical-shift filtering. J Am Chem Soc 124:13938–13948
Traaseth NJ, Veglia G (2011) Frequency-selective heteronuclear dephasing and selective carbonyl labeling to deconvolute crowded spectra of membrane proteins by magic angle spinning NMR. J Magn Reson 211:18–24
Tycko R (2007) Stochastic dipolar recoupling in nuclear magnetic resonance of solids. Phys Rev Lett 99:187601
Ulrich EL et al (2008) Bio Mag Res Bank. Nucleic Acids Res 36:D402–D408
Verel R, Baldus M, Ernst M, Meier BH (1998) A homonuclear spin-pair filter for solid-state NMR based on adiabatic-passage techniques. Chem Phys Lett 287:421–428
Verel R, Ernst M, Meier BH (2001) Adiabatic dipolar recoupling in solid-state NMR: The DREAM scheme. J Magn Reson 150:81–99
Westfeld T, Verel R, Ernst M, Bockmann A, Meier BH (2012) Properties of the DREAM scheme and its optimization for application to proteins. J Biomol NMR 53:103–112
Williams JK, Schmidt-Rohr K, Hong M (2015) Aromatic spectral editing techniques for magic-angle-spinning solid-state NMR spectroscopy of uniformly C-13-labeled proteins. Solid State Nucl Magn Reson 72:118–126
Wu XL, Burns ST, Zilm KW (1994) Spectral editing in CPMAS NMR - generating subspectra based on proton multiplicities. J Magn Reson, Ser A 111:29–36
Wu XL, Zilm KW (1993) Complete spectral editing in CPMAS NMR. J Magn Reson, Ser A 102:205–213
Xie H, Zhao Y, Wang J, Zhang Z, Yang J (2018) Solid-state NMR chemical shift assignments of aquaporin Z in lipid bilayers. Biomol NMR Assign 12:323–328
Zech SG, Olejniczak E, Hajduk P, Mack J, McDermot AE (2004) Characterization of protein-ligand interactions by high-resolution solid-state NMR spectroscopy. J Am Chem Soc 126:13948–13953
Zhang ZF, Liu H, Deng J, Tycko R, Yang J (2019) Optimization of band-selective homonuclear dipolar recoupling in solid-state NMR by a numerical phase search. J Chem Phys 150:154201
Zhang ZF et al (2020) Selectively Enhanced H-1-H-1 Correlations in Proton-Detected Solid-State NMR under Ultrafast MAS Conditions. J Phys Chem Lett 11:8077–8083
Zhao Y et al (2018) Gating mechanism of aquaporin Z in synthetic bilayers and native membranes revealed by solid-state NMR spectroscopy. J Am Chem Soc 140:7885–7895
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2016YFA0501200, 2017YFA0505400), the National Natural Science Foundation of China (21775161, 22074153, 21927801, 31627803, 31770798 and 21921004) and Chinese Academy of Sciences (YJKYYQ20190032).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, H., Zhang, Z., Zhao, Y. et al. Spectral editing of alanine, serine, and threonine in uniformly labeled proteins based on frequency-selective homonuclear recoupling in solid-state NMR. J Biomol NMR 75, 193–202 (2021). https://doi.org/10.1007/s10858-021-00367-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-021-00367-9