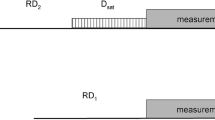
Abstract
Spectral resolution remains one of the most significant limitations in the NMR study of biomolecules. We present the srNOESY (super resolution nuclear overhauser effect spectroscopy) experiment, which enhances the resolution of NOESY cross-peaks at the expense of the diagonal peak line-width. We studied two proteins, ubiquitin and the influenza hemagglutinin fusion peptide in bicelles, and we achieved average resolution enhancements of 21–47% and individual peak enhancements as large as ca. 450%. New peaks were observed over the conventional NOESY experiment in both proteins as a result of these improvements, and the final structures generated from the calculated restraints matched published models. We discuss the impact of the experimental parameters, spin diffusion and the information content of the srNOESY lineshape.










Similar content being viewed by others
References
Baleja JD, Moult J, Sykes BD (1990) Distance measurement and structure refinement with NOE data. J Magn Reson 87:375–384
Battiste JL, Wagner G (2000) Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry 39:5355–5365
Borgias BA, Gochin M, Kerwood DJ, James TL (1990) Relaxation matrix analysis of 2D NMR data. Prog Nucl Magn Reson Spectrosc 22:83–100
Cavanagh J, Fairbrother W, Palmer III, Rance M, Skelton N (2007) Protein NMR principles and practice. Academic Press, Cambridge. https://doi.org/10.1093/cid/cis040
Chi CN, Strotz D, Riek R, Vögeli B (2018) NOE-derived methyl distances from a 360 kDa PROTEASOME Complex. Chemistry 24:2270–2276
Cole R, Loria JP, FAST-Modelfree (2003) A program for rapid automated analysis of solution NMR spin-relaxation data. J Biomol NMR 26:203–213
Cornilescu G, Marquardt JL, Ottiger M, Bax A (1998) Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J Am Chem Soc 120:6836–6837
Delaglio F et al (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Dobson C, Olejniczak E, Poulsen F, Ratcliffe R (1982) Time development of proton nuclear overhauser effects in proteins. J Magn Reson 48:97–110
Ellman JA, Volkman BF, Mendel D, Schulz PG, Wemmer DE (1992) Site-specific isotopic labeling of proteins for NMR studies. J Am Chem Soc 114:7959–7961
Farrow NA et al (1994) Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by15N NMR relaxation. Biochemistry 33:5984–6003
Goddard TD, Kneller DG (2008) SPARKY 3. University of California, San Fransisco
Kalbitzer HR, Leberman R, Wittinghofer A (1985) 1H-NMR spectroscopy on elongation factor Tu from Escherichia coli: resolution enhancement by perdeuteration. FEBS Lett 180:40–42
Kay LE, Keifer P, Saarinen T (1992) Pure absorption gradient enhanced heteronucler single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc 114:10663–10665
Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327
LeMaster DM (1989) Deuteration in protein proton magnetic resonance. Methods Enzymol 177:23–43
LeMaster DM, Richards FM (1988) NMR sequential assignment of Escherichia coli thioredoxin utilizing random fractional deuteriation. Biochemistry 27:142–150
Lipari G, Szabo A (1982a) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc 104:4559–4570
Lipari G, Szabo A (1982b) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc 104:4546–4559
Lorieau JL, Louis JM, Bax A The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc Natl Acad Sci 107:11341–11346 (2010)
Maciejewski MW et al (2017) NMRbox: a resource for biomolecular NMR computation. Biophys J 112:1529–1534
Macura S, Wüthrich K, Ernst RR (1982a) The relevance of J cross-peaks in two-dimensional NOE experiments of macromolecules. J Magn Reson 47:351–357
Macura S, Wuthrich K, Ernst RR (1982b) Separation and suppression of coherent transfer effects in two-dimensional NOE and chemical exchange spectroscopy. J Magn Reson 46:269–282
Marley J, Lu M, Bracken C (2001) A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR 20:71–75
Muhandiram R, Kay LE (2007) Three-Dimensional HMQC-NOESY, NOESY-HMQC, and NOESY-HSQC. Encycl Magn Reson. https://doi.org/10.1002/9780470034590.emrstm0563
Nelson DL, Cox MM (2013) Lehninger principles of biochemistry. W. H. Freeman and Company, New York
Neuhaus D, Williamson MP (2000) The nuclear overhauser effect in structural and conformational analysis. Wiley, New Jersey. https://doi.org/10.1016/0022-2364(92)90256-7
Ollerenshaw JE, Tugarinov V, Kay LE, Methyl TROSY (2003) Explanation and experimental verification. Magn Reson Chem 41:843–852
Otten R, Chu B, Krewulak KD, Vogel HJ, Mulder FAA (2010) Comprehensive and cost-effective NMR spectroscopy of methyl groups in large proteins. J Am Chem Soc 132:2952–2960
Pervushin K, Riek R, Wider G, Wuthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci 94:12366–12371
Rieping W, Bardiaux B, Bernard A, Malliavin TE, Nilges M (2007) ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 23:381–382
Schimmel PR, Cantor CR (1980) Biophysical chemistry, part II: techniques for the study of biological structure and function. W.H. Freeman, San Francisco
Schwieters CD, Kuszewski JJ Clore GM (2006) Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc 48:47–62
Shaka AJ, Keeler J, Frenkiel T, Freeman R (1983) An improved sequence for broadband decoupling: WALTZ-16. J Magn Reson 52:335–338
Smrt ST, Draney AW, Lorieau JL (2015) The influenza hemagglutinin fusion domain is an amphipathic helical hairpin that functions by inducing membrane curvature. J Biol Chem 290:228–238
Tugarinov V, Kay LE (2005) Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. ChemBioChem 6:1567–1577
Vögeli B et al (2009) Exact distances and internal dynamics of perdeuterated ubiquitin from NOE buildups. J Am Chem Soc 131:17215–17225
Vögeli B, Friedmann M, Leitz D, Sobol A, Riek R (2010) Quantitative determination of NOE rates in perdeuterated and protonated proteins: Practical and theoretical aspects. J Magn Reson 204:290–302
Vuister GW, Bax A (1992) Resolution enhancement and spectral editing of uniformly 13C-enriched proteins by homonuclear broadband 13C decoupling. J Magn Reson 98:428–435
Weigelt J (1998) Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J Am Chem Soc 120:10778–10779
Acknowledgements
This work was supported by the National Science Foundation under Grant No. 1651598 and funds from the Department of Chemistry at the University of Illinois at Chicago. This study made use of NMRbox: National Center for Biomolecular NMR Data Processing and Analysis, a Biomedical Technology Research Resource (BTRR), which is supported by NIH Grant P41GM111135 (NIGMS).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
DeLisle, C.F., Mendis, H.B. & Lorieau, J.L. Super resolution NOESY spectra of proteins. J Biomol NMR 73, 105–116 (2019). https://doi.org/10.1007/s10858-019-00231-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-019-00231-x