Abstract
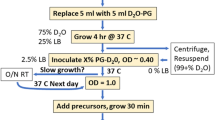
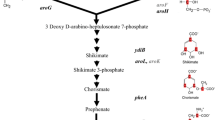
The amino acids 4-(tert-butyl)phenylalanine (Tbf) and 4-(trimethylsilyl)phenylalanine (TMSf), as well as a partially deuterated version of Tbf (dTbf), were chemically synthesized and site-specifically incorporated into different proteins, using an amber stop codon, suppressor tRNA and the broadband aminoacyl-tRNA synthetase originally evolved for the incorporation of p-cyano-phenylalanine. The 1H-NMR signals of the tert-butyl and TMS groups were compared to the 1H-NMR signal of tert-butyltyrosine (Tby) in protein systems with molecular weights ranging from 8 to 54 kDa. The 1H-NMR resonance of the TMS group appeared near 0 ppm in a spectral region with few protein resonances, facilitating the observation of signal changes in response to ligand binding. In all proteins, the R 2 relaxation rate of the tert-butyl group of Tbf was only little greater than that of Tby (less than two-fold). Deuteration of the phenyl ring of Tbf made only a relatively small difference. The effective T 2 relaxation time of the TMS signal was longer than 140 ms even in the 54 kDa system.



Similar content being viewed by others
References
Apponyi MA, Ozawa K, Dixon NE, Otting G (2008) Cell-free protein synthesis for analysis by NMR spectroscopy. In: Kobe B, Guss M, Huber T (eds) Structural proteomics: high-throughput methods. Humana Press, Totowa, pp 257–268
Chen W-N, Kuppan KV, Lee MD, Jaudzems K, Huber T, Otting G (2015) O-tert-butyltyrosine, an NMR tag for high-molecular-weight systems and measurements of submicromolar ligand binding affinities. J Am Chem Soc 137:4581–4586
Chong S (2001) Overview of cell-free protein synthesis: historic landmarks, commercial systems, and expanding applications. In Current protocols in molecular biology. Wiley, New York
Gan Q, Fan C (2017) Increasing the fidelity of noncanonical amino acid incorporation in cell-free protein synthesis. Biochim Biophys Acta 1861:3047–3052
George S, Aguirre JD, Spratt DE, Bi Y, Jeffery M, Shaw GS, O’Donoghue P (2016) Generation of phospho-ubiquitin variants by orthogonal translation reveals codon skipping. FEBS Lett 590:1530–1542
Hammill JT, Miyake-Stoner S, Hazen JL, Jackson JC, Mehl RA (2007) Preparation of site-specifically labeled fluorinated proteins for 19F-NMR structural characterization. Nat Protoc 2:2601–2607
Hubbard PS (1958) Nuclear magnetic relaxation of three and four spin molecules in a liquid. Phys Rev 109:1153–1158
Hwang T-L, Shaka AJ (1995) Water suppression that works-excitation sculpting using arbitrary waveforms and pulsed field gradients. J Magn Reson A 112:275–279
Jabar S, Adams LA, Wang Y, Aurelio L, Graham B, Otting G (2017) Chemical tagging with tert-butyl and trimethylsilyl groups for measuring intermolecular nuclear Overhauser effects in a large protein–ligand complex. Chem Eur J 23:13033–13036
Loscha KV, Herlt AJ, Qi R, Huber T, Ozawa K, Otting G (2012) Multiple-site labeling of proteins with unnatural amino acids. Angew Chem Int Ed 51:2243–2246
Mahawaththa MC, Pearce BJG, Szabo M, Graham B, Klein CD, Nitsche C, Otting G (2017) Solution conformations of a linked construct of the Zika virus NS2B-NS3 protease. Antiviral Res 142:141–147
Mukai T, Hayashi A, Iraha F, Sato A, Ohtake K, Yokoyama S, Sakamoto K (2010) Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res 38:8188–8195
Mukai T, Hoshi H, Ohtake K, Takahashi M, Yamaguchi A, Hayashi A, Yokoyama S, Sakamoto K (2015) Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci Rep 5:9699
Müller N, Bodenhausen G, Ernst RR (1987) Relaxation-induced violations of coherence transfer selection rules in nuclear magnetic resonance. J Magn Reson 75:297–334
Neylon C, Brown SE, Kralicek AV, Miles CS, Love CA, Dixon NE (2000) Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: a model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry 39:11989–11999
Nilsson M, Rydén-Aulin M (2003) Glutamine is incorporated at the nonsense codons UAG and UAA in a suppressor-free Escherichia coli strain. Biochim Biophys Acta 1627:1–6
O’Donoghue P, Prat L, Heinemann IU, Ling J, Odoi K, Liu WR, Söll D (2012) Near-cognate suppression of amber, opal and quadruplet codons competes with aminoacyl-tRNAPyl for genetic code expansion. FEBS Lett 586:3931–3937
Ohtake K, Sato A, Mukai T, Hino N, Yokoyama S, Sakamoto K (2012) Efficient decoding of the UAG triplet as a full-fledged sense codon enhances the growth of a prfA-deficient strain of Escherichia coli. J Bacteriol 194:2606–2613
Ozawa K, Loscha KV, Kuppan KV, Loh CT, Dixon NE, Otting G (2012) High-yield cell-free protein synthesis for site-specific incorporation of unnatural amino acids at two sites. Biochem Biophys Res Commun 418:652–656
Torizawa T, Shimizu M, Taoka M, Miyano H, Kainosho M (2004) Efficient production of isotopically labeled proteins by cell-free synthesis: a practical protocol. J Biomol NMR 30:311–325
Werbelow LG, Marshall AG (1973) Internal rotation and nonexponential methyl nuclear relaxation for macromolecules. J Magn Reson 11:299–313
Young TS, Ahmad I, Yin JA, Schultz PG (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol 395:361–374
Young DD, Young TS, Jahnz M, Ahmad I, Spraggon G, Schultz PG (2011) An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 50:1894–1900
Acknowledgements
We thank Mr Alireza Bahramzadeh for an expression construct and sample of wild-type Leu-RS, Dr Yao Wang and Mr Mithun Chamikara Mahawaththa for mass spectrometry measurements, and Professor Peter G. Schultz for the pEvol-pCNF-RS plasmid. Financial support by the Australian Research Council is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Loh, C.T., Adams, L.A., Graham, B. et al. Genetically encoded amino acids with tert-butyl and trimethylsilyl groups for site-selective studies of proteins by NMR spectroscopy. J Biomol NMR 71, 287–293 (2018). https://doi.org/10.1007/s10858-017-0157-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-017-0157-y