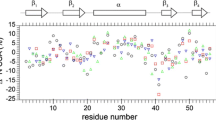
Abstract
Description of protein dynamics is known to be essential in understanding their function. Studies based on a well established \(^{15}\hbox {N}\) NMR relaxation methodology have been applied to a large number of systems. However, the low dispersion of \(^{1}\hbox {H}\) chemical shifts very often observed within intrinsically disordered proteins complicates utilization of standard 2D HN correlated spectra because a limited number of amino acids can be characterized. Here we present a suite of triple resonance HNCO-type NMR experiments for measurements of five \(^{15}\hbox {N}\) relaxation parameters (\(R_1\), \(R_2\), NOE, cross-correlated relaxation rates \(\Gamma _x\) and \(\Gamma _z\)) in doubly \(^{13}\hbox {C}\),\(^{15}\hbox {N}\)-labeled proteins. We show that the third spectral dimension combined with non-uniform sampling provides relaxation rates for almost all residues of a protein with extremely poor chemical shift dispersion, the C terminal domain of \(\delta\)-subunit of RNA polymerase from Bacillus subtilis. Comparison with data obtained using a sample labeled by \(^{15}\hbox {N}\) only showed that the presence of \(^{13}\hbox {C}\) has a negligible effect on \(\Gamma _x\), \(\Gamma _z\), and on the cross-relaxation rate (calculated from NOE and \(R_1\)), and that these relaxation rates can be used to calculate accurate spectral density values. Partially \(^{13}\hbox {C}\)-labeled sample was used to test if the observed increase of \(^{15}\hbox {N}\) \(R_1\) in the presence of \(^{13}\hbox {C}\) corresponds to the \(^{15}\hbox {N}-^{13}\hbox {C}\) dipole–dipole interactions in the \(^{13}\hbox {C}\),\(^{15}\hbox {N}\)-labeled sample.










Similar content being viewed by others
References
Bertoncini C (2007) Structural characterization of the intrinsically unfolded protein \(\beta\)-synuclein, a natural negative regulator of \(\alpha\)-synuclein aggregation. J Mol Biol 372:708–722
Burnley B, Kalverda A, Paisey S, Berry A, Homans S (2007) Hadamard NMR spectroscopy for relaxation measurements of large (\(\ge\)35 kDa) proteins. J Biomol NMR 39:239–245
Caffrey M (1998) 3D NMR experiments for measuring 15N relaxation data of large proteins: application to the 44 kDa ectodomain of SIV gp41. J Magn Reson 135:368–372
Carr P, Fearing D, Palmer A III (1998) 3D accordion spectroscopy for measuring 15N and 13CO relaxation rates in poorly resolved NMR spectra. J Magn Reson 132:25–33
Chill JH, Louis JM, Baber JL, Bax A (2006) Measurement of \(^{15}\)N relaxation in the detergent-solubilized tetrameric KcsA potassium channel. J Biomol NMR 36:123–136
Csizmok V, Felli I, Tompa P, Banci L, Bertini I (2008) Structural and dynamic characterization of intrinsically disordered human securin by NMR spectroscopy. J Am Chem Soc 130:16873–16879
d’Auvergne E, Gooley P (2008) Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J Biomol NMR 40:107–119
d’Auvergne E, Gooley P (2008) Optimisation of NMR dynamic models II. A new methodology for the dual optimisation of the model-free parameters and the Brownian rotational diffusion tensor. J Biomol NMR 40:121–133
Delaglio F (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Demo G et al (2014) X-ray vs. NMR structure of N-terminal domain of \(\delta\)-subunit of RNA polymerase. J Struct Biol 187:174–186
Dunker A, Obradovic Z, Romero P, Garner E, Brown C (2000) Intrinsic protein disorder in complete genomes. Genome Inf 11:161–171
Dyson HJ (2011) Expanding the proteome: disordered and alternatively folded proteins. Q Rev Biophys 44:467–518
Dyson H, Wright P (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208
Eaton JW, Bateman D, Hauberg S, Wehbring R (2016) GNU Octave version 4.2.0 manual: a high-level interactive language for numerical computations
Efron B (1979) Bootstrap methods: another look at the jackknife. Ann Stat 7:1–26
Eliezer D (2012) Distance information for disordered proteins from NMR and ESR measurements using paramagnetic spin labels. Methods Mol Biol 895:127–138
Engh R, Huber R (1991) Accurate bond and angle parameters for Xray protein structure refinement. Acta Cryst A 47:392–400
Farrow NA, Zhang O, Szabo A, Torchia DA, Kay LE (1995) Spectral density function mapping using \(^{15}\)N relaxation data exclusively. J Biomol NMR 6:153–162
Ferrage F (2012) Protein dynamics by \(^{15}\)N nuclear magnetic relaxation. Methods Mol Biol 831:141–163
Ferrage F, Pelupessy P, Cowburn D, Bodenhausen G (2006) Protein backbone dynamics through \(^{13}\)C-\(^{13}\)C\(^{\alpha }\) cross-relaxation in NMR spectroscopy. J Am Chem Soc 128:11072–11078
Ferrage F, Reichel A, Battacharya S, Cowburn D, Ghose R (2010) On the measurement of \(^{15}\)N-\(^1\)H nuclear Overhauser effects. 2. Effects of the saturation scheme and water signal suppression. J Magn Reson 207:294–303
Ghose R, Huang K, Prestegard J (1998) Measurement of cross correlation between dipolar coupling and chemical shift anisotropy in the spin relaxation of \(^{13}\)C, \(^{15}\)N-labeled proteins. J Magn Reson 135:487–499
Goddard TD, Kneller DG (2003) Sparky 3. University of California, San Francisco
Grzesiek S, Bax A (1993) Amino acid type determination in the sequential assignment procedure of uniformly \(^{13}\)C/ \(^{15}\)N-enriched proteins. J Biomol NMR 3:185–204
Hall J, Fushman D (2003) Direct measurement of the transverse and longitudinal \(^{15}\)N chemical shift anisotropy-dipolar cross-correlation rate constants using \(^{1}\)H-coupled HSQC spectra. Magn Reson Chem 41:837–842
Jarymowycz V, Stone M (2006) Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem Rev 106:1624–1671
Kadeřávek P (2014) Spectral density mapping protocols for analysis of molecular motions in disordered proteins. J Biomol NMR 58:193–207
Kay L, Torchia D, Bax A (1989) Backbone dynamics of proteins as studied by \(^{15}\)N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28:8972–8979
Kay LE, Ikura M, Tschudin R, Bax A (1990) Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J Magn Reson 213:423–441
Kay L, Xu G, Yamazaki T (1994) Enhanced-sensitivity triple-resonance spectroscopy with minimal H\(_{2}\)O saturation. J Magn Reson A 109:129–133
Kazimierczuk K, Zawadzka A, Koźmiński W, Zhukov I (2006) Random sampling of evolution time space and Fourier transform processing. J Biomol NMR 36:157–168
Kazimierczuk K, Zawadzka A, Koźmiński W, Zhukov I (2007) Lineshapes and artifacts in Multidimensional Fourier Transform of arbitrary sampled NMR data sets. J Magn Reson 188:344–356
Kneller JM, Lu M, Bracken C (2002) An effective method for the discrimination of motional anisotropy and chemical exchange. J Am Chem Soc 124:1852–1853
Korzhnev D, Billeter M, Arseniev A, Orekhov V (2001) NMR studies of Brownian tumbling and internal motions in proteins. Prog Nucl Magn Reson Spectrosc 38:197–266
Krishna N, Berliner L (2006) Structure computation and dynamics in protein NMR. Biological magnetic resonance. New York, Springer
Lakomek N-A, Kaufman JD, Stahl SJ, Louis JM, Grishaev A, Wingfield PT, Bax A (2013) Internal dynamics of the homotrimeric HIV-1 viral coat protein gp41 on multiple time scales. Angew Chem Int Ed Engl 52:3911–3915
Lakomek N-A, Ying J, Bax A (2012) Measurement of \(^{15}\)N relaxation rates in perdeuterated proteins by TROSY-based methods. J Biomol NMR 53:209–221
Lawrence CW, Showalter SA (2012) Carbon-detected \(^{15}\)N NMR spin relaxation of an intrinsically disordered protein: FCP1 dynamics unbound and in complex with RAP74. J Phys Chem Lett 3:1409–1413
Libich D, Harauz G (2008) Backbone dynamics of the 18.5 kDa isoform of myelin basic protein reveals transient \(\alpha\)-helices and a calmodulin-binding site. Biophys J 94:4847–4866
Liu J (2006) Intrinsic disorder in transcription factors. Biochemistry 45:6873–6888
Lopez de Saro F, Moon Woody A-Y, Helmann J (1995) Structural analysis of the Bacillus subtilis \(\delta\) factor: a protein polyanion which displaces RNA from RNA polymerase. J Mol Biol 252:189–202
Motáčková V et al (2010) Strategy for complete NMR assignment of disordered proteins with highly repetitive sequences based on resolution-enhanced 5D experiments. J Biomol NMR 48:169–177
Niedermeyer THJ, Strohalm M (2012) mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS ONE 7:e44913
Nováček J, Janda L, Dopitová R, Žídek L, Sklenář V (2013) Efficient protocol for backbone and side-chain assignments of large, intrinsically disordered proteins: transient secondary structure analysis of 49.2 kda microtubule associated protein 2c. J Biomol NMR 56:291–301
Nováček J, Žídek L, Sklenář V (2014) Toward optimal-resolution NMR of intrinsically disordered proteins. J Magn Reson 241:41–52
Olson K (2005) Secondary structure and dynamics of an intrinsically unstructured linker domain. J Biomol Struct Dyn 23:113–124
Papoušková V (2013) Structural study of the partially disordered full-length \(\delta\)-subunit of RNA polymerase from Bacillus subtilis. ChemBioChem 14:1772–1779
Pelupessy P, Espallargas G, Bodenhausen G (2003) Symmetrical reconversion: measuring cross-correlation rates with enhanced accuracy. J Magn Reson 161:258–264
Pelupessy P, Ferrage F, Bodenhausen G (2007) Accurate measurement of longitudinal cross-relaxation rates in nuclear magnetic resonance. J Chem Phys 126:134508
Powers R, Gronenborn A, Marius Clore G, Bax A (1991) Three-dimensional triple-resonance NMR of \(^{13}\)C/\(^{15}\)N-enriched proteins using constant-time evolution. J Magn Reson (1969) 94:209–213
Schleucher J, Sattler M, Griesinger C (1993) Coherence selection by gradients without signal attenuation: application to the three-dimensional HNCO experiment. Angew Chem Int Ed 32:1489–1491
Sibille N, Bernad P (2012) Structural characterization of intrinsically disordered proteins by the combined use of NMR and SAXS. Biochem Soc Trans 40:955–962
Stadler A (2014) Internal nanosecond dynamics in the intrinsically disordered myelin basic protein. J Am Chem Soc 136:6987–6994
Stanek J, Koźmiński W (2010) Iterative algorithm of discrete Fourier transform for processing randomly sampled NMR data sets. J Biomol NMR 47:65–77
Stanek J, Saxena S, Geist L, Konrat R, Koźmiński W (2013) Probing local backbone geometries in intrinsically disordered proteins by cross-correlated NMR relaxation. Angew Chem Int Ed 52:4604–4606
Strohalm M, Hassman M, Košata B, Kodíček M (2008) mMass data miner: an open source alternative for mass spectrometric data analysis. Rapid Commun Mass Spectrom 22:905–908
Strohalm M, Kavan D, Novák P, Volný M, Havlíček V (2010) mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal Chem 82:4648–4651
Sykes MT, Williamson JR (2008) Envelope: interactive software for modeling and fitting complex isotope distributions. BMC Bioinf 9:446
Tjandra N, Feller S, Pastor R, Bax A (1995) Rotational diffusion anisotropy of human ubiquitin from \(^{15}\)N NMR relaxation. J Am Chem Soc 117:12562–12566
Tompa P (2005) The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett 579:3346–3354
Tompa K, Bokor M, Tompa P (2012) Wide-line NMR and protein hydration. Methods Mol Biol 895:167–196
Tugarinov V, Choy W, Kupce E, Kay L (2004) Addressing the overlap problem in the quantitative analysis of two dimensional NMR spectra: application to \(^{15}\)N relaxation measurements. J Biomol NMR 30:347–352
Uversky VN (2013) The most important thing is the tail: multitudinous functionalities of intrinsically disordered protein termini. FEBS Lett 587:1891–1901
Viles JH (2001) Local structural plasticity of the prion protein. Analysis of NMR relaxation dynamics. Biochemistry 40:2743–2753
Vise P, Baral B, Latos A, Daughdrill G (2005) NMR chemical shift and relaxation measurements provide evidence for the coupled folding and binding of the p53 transactivation domain. Nucleic Acids Res 33:2061–2077
Acknowledgements
This work was supported by Czech Science Foundation, Grant Number GA 13-16842S and by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR) under the project CEITEC 2020 (LQ1601). P.S. was supported by project “Employment of Newly Graduated Doctors of Science for Scientific Excellence” (Grant Number CZ.1.07/2.3.00/30.0009) from the European Social Fund and the state budget of the Czech Republic and partially by MEYS CR (project LO1304). J. B. received Institutional support RVO:68081715 from the Institute of Analytical Chemistry of the Czech Academy of Sciences, v.v.i. CIISB research infrastructure project LM2015043 funded by MEYS CR is gratefully acknowledged for a partial financial support of the measurements at the Josef Dadok National NMR Centre, CEITEC - Masaryk University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Srb, P., Nováček, J., Kadeřávek, P. et al. Triple resonance 15N NMR relaxation experiments for studies of intrinsically disordered proteins. J Biomol NMR 69, 133–146 (2017). https://doi.org/10.1007/s10858-017-0138-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-017-0138-1