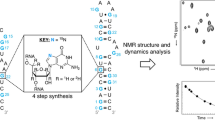
Abstract
In comparison to proteins and protein complexes, the size of RNA amenable to NMR studies is limited despite the development of new isotopic labeling strategies including deuteration and ligation of differentially labeled RNAs. Due to the restricted chemical shift dispersion in only four different nucleotides spectral resolution remains limited in larger RNAs. Labeling RNAs with the NMR-active nucleus 19F has previously been introduced for small RNAs up to 40 nucleotides (nt). In the presented work, we study the natural occurring RNA aptamer domain of the guanine-sensing riboswitch comprising 73 nucleotides from Bacillus subtilis. The work includes protocols for improved in vitro transcription of 2-fluoroadenosine-5′-triphosphat (2F-ATP) using the mutant P266L of the T7 RNA polymerase. Our NMR analysis shows that the secondary and tertiary structure of the riboswitch is fully maintained and that the specific binding of the cognate ligand hypoxanthine is not impaired by the introduction of the 19F isotope. The thermal stability of the 19F-labeled riboswitch is not altered compared to the unmodified sequence, but local base pair stabilities, as measured by hydrogen exchange experiments, are modulated. The characteristic change in the chemical shift of the imino resonances detected in a 1H,15N-HSQC allow the identification of Watson–Crick base paired uridine signals and the 19F resonances can be used as reporters for tertiary and secondary structure transitions, confirming the potential of 19F-labeling even for sizeable RNAs in the range of 70 nucleotides.







Similar content being viewed by others
References
Alvarado LJ et al (2014) Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates RNA structure and dynamics studies. ChemBioChem 15:1573–1577. doi:10.1002/cbic.201402130
Appasamy D, Ramlan EI, Firdaus-Raih M (2013) Comparative sequence and structure analysis reveals the conservation and diversity of nucleotide positions and their associated tertiary interactions in the riboswitches. PLoS One. doi:10.1371/journal.pone.0073984
Baldo JH, Hansen PE, Shriver JW, Sykes BD (1983) 2-Fluoro-Atp, a fluorinated Atp Analog: F-19 nuclear magnetic-resonance studies of the 2-fluoro-Adp. Myosin subfragment-1 complex. Can J Biochem Cell B 61:115–119
Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR (1992) Preparation of isotopically labeled ribonucleotides for multidimensional NMR-spectroscopy of RNA. Nucleic Acids Res 20:4515–4523. doi:10.1093/nar/20.17.4515
Batey RT, Gilbert SD, Montange RK (2004) Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432:411–415. doi:10.1038/nature03037
Battaglia MR, Buckingham AD, Williams JH (1981) The electric quadrupole-moments of benzene and hexafluorobenzene. Chem Phys Lett 78:420–423
Broom AD, Amarnath V, Vince R, Brownell J (1979) Poly(2-fluoroadenylic acid). The role of basicity in the stabilization of complementary helices. Biochim Biophys Acta 563:508–517
Buck J, Furtig B, Noeske J, Wohnert J, Schwalbe H (2007) Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc Natl Acad Sci USA 104:15699–15704. doi:10.1073/pnas.0703182104
Buck J, Noeske J, Wohnert J, Schwalbe H (2010) Dissecting the influence of Mg2+ on 3D architecture and ligand-binding of the guanine-sensing riboswitch aptamer domain. Nucleic Acids Res 38:4143–4153. doi:10.1093/nar/gkq138
Butcher SE, Allain FHT, Feigon J (2000) Determination of metal ion binding sites within the hairpin ribozyme domains by NMR. Biochemistry 39:2174–2182. doi:10.1021/Bi9923454
Carlomagno T, Amata I, Codutti L, Falb M, Fohrer J, Masiewicz P, Simon B (2013) Structural principles of RNA catalysis in a 2′–5′ lariat-forming ribozyme. J Am Chem Soc 135:4403–4411. doi:10.1021/Ja311868t
Cavaluzzi MJ, Borer PN (2004) Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. doi:10.1093/nar/gnh015
Chowdhury S, Maris C, Allain FHT, Narberhaus F (2006) Molecular basis for temperature sensing by an RNA thermometer. EMBO J 25:2487–2497. doi:10.1038/sj.emboj.7601128
Christiansen LC, Schou S, Nygaard P, Saxild HH (1997) Xanthine metabolism in Bacillus subtilis: characterization of the xpt–pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J Bacteriol 179:2540–2550
Codington JF, Fox JJ, Doerr IL (1964) Nucleosides. 18. Synthesis of 2]-fluorothymidine 2]-fluorodeoxyuridine + other 2]-halogeno-2]-deoxy nucleosides. J Org Chem 29:558–564. doi:10.1021/Jo01026a009
Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE (2005) RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J Mol Biol 351:371–382. doi:10.1016/j.jmb.2005.05.069
Dayie KT, Tolbert TJ, Williamson JR (1998) 3D C(CC)H TOCSY experiment for assigning protons and carbons in uniformly C-13- and selectively H-2-labeled RNA. J Magn Reson 130:97–101. doi:10.1006/jmre.1997.1286
D’Souza V, Dey A, Habib D, Summers MF (2004) NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J Mol Biol 337:427–442. doi:10.1015/j.jmb.2004.01.037
Duszczyk MM, Wutz A, Rybin V, Sattler M (2011) The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 17:1973–1982. doi:10.1261/Rna.2747411
Fauster K, Kreutz C, Micura R (2012) 2′-SCF3 uridine-a powerful label for probing structure and function of RNA by 19F NMR spectroscopy. Angew Chem Int Ed Engl 51:13080–13084. doi:10.1002/anie.201207128
Ferner J et al (2009) Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries. ChemBioChem 10:1490–1494. doi:10.1002/cbic.200900220
Furtig B, Richter C, Wohnert J, Schwalbe H (2003) NMR spectroscopy of RNA. Chembiochem 4:936–962. doi:10.1002/cbic.200300700
Furtig B, Richter C, Schell P, Wenter P, Pitsch S, Schwalbe H (2008) NMR-spectroscopic characterization of phosphodiester bond cleavage catalyzed by the minimal hammerhead ribozyme. RNA Biol 5:41–48
Garavis M et al (2014) Discovery of selective ligands for telomeric RNA G-quadruplexes (TERRA) through 19F-NMR based fragment screening. ACS Chem Biol 9:1559–1566. doi:10.1021/cb500100z
Geen H, Freeman R (1991) Band-selective radiofrequency pulses. J Magn Reson 93:93–141. doi:10.1016/0022-2364(91)90034-Q
Graber D, Moroder H, Micura R (2008) 19F NMR spectroscopy for the analysis of RNA secondary structure populations. J Am Chem Soc 130:17230–17231. doi:10.1021/ja806716s
Granqvist L, Virta P (2014) 4′-C-[(4-trifluoromethyl-1H-1,2,3-triazol-1-yl)methyl]thymidine as a sensitive (19)F NMR sensor for the detection of oligonucleotide secondary structures. J Org Chem 79:3529–3536. doi:10.1021/jo500326j
Guillerez J, Lopez PJ, Proux F, Launay H, Dreyfus M (2005) A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc Natl Acad Sci USA 102:5958–5963. doi:10.1073/pnas.0407141102
Horowitz J, Ou CN, Ishaq M (1974) Isolation and partial characterization of Escherichia coli valine transfer RNA with uridine-derived residues replaced by 5-fluorouridine. J Mol Biol 88:301–312
Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D’Souza VM (2011) An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480:561–564. doi:10.1038/Nature10657
Huang P, Plunkett W (1987) Phosphorolytic cleavage of 2-fluoroadenine from 9-beta-D-arabinofuranosyl-2-fluoroadenine by Escherichia-Coli: a pathway for 2-fluoro-Atp production. Biochem Pharmacol 36:2945–2950. doi:10.1016/0006-2952(87)90207-3
Keane SC et al (2015) Structure of the HIV-1 RNA packaging signal. Science 348:917–921. doi:10.1126/science.aaa9266
Kiviniemi A, Virta P (2010) Characterization of RNA invasion by (19)F NMR spectroscopy. J Am Chem Soc 132:8560–8562. doi:10.1021/ja1014629
Kiviniemi A, Virta P (2011) Synthesis of aminoglycoside-3′-conjugates of 2′-O-methyl oligoribonucleotides and their invasion to a 19F labeled HIV-1 TAR model. Bioconjug Chem 22:1559–1566. doi:10.1021/bc200101r
Kiviniemi A, Murtola M, Ingman P, Virta P (2013) Synthesis of fluorine-labeled peptide nucleic acid building blocks as sensors for the 19F NMR spectroscopic detection of different hybridization modes. J Org Chem 78:5153–5159. doi:10.1021/jo400014y
Kline PC, Serianni AS (1990) C-13-Enriched ribonucleosides: synthesis and application of C-13-H-1 and C-13-C-13 spin-coupling constants to assess furanose and N-glycoside bond conformations. J Am Chem Soc 112:7373–7381. doi:10.1021/Ja00176a043
Kosutic M et al (2014) Surprising base pairing and structural properties of 2′-trifluoromethylthio-modified ribonucleic acids. J Am Chem Soc 136:6656–6663. doi:10.1021/ja5005637
Kreutz C, Kahlig H, Konrat R, Micura R (2005) Ribose 2′-F labeling: a simple tool for the characterization of RNA secondary structure equilibria by 19F NMR spectroscopy. J Am Chem Soc 127:11558–11559. doi:10.1021/ja052844u
Kreutz C, Kahlig H, Konrat R, Micura R (2006) A general approach for the identification of site-specific RNA binders by 19F NMR spectroscopy: proof of concept. Angew Chem Int Ed Engl 45:3450–3453. doi:10.1002/anie.200504174
Lescoute A, Westhof E (2006) The A-minor motifs in the decoding recognition process. Biochimie 88:993–999. doi:10.1016/j.biochi.2006.05.018
Levitt MH, Freeman R (1981) Composite pulse decoupling. J Magn Reson 43:502–507. doi:10.1016/0022-2364(81)90066-4
Lombes T et al (2012) Investigation of RNA-ligand interactions by 19F NMR spectroscopy using fluorinated probes. Angew Chem Int Ed Engl 51:9530–9534. doi:10.1002/anie.201204083
Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR (2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113:577–586
Marchanka A, Simon B, Althoff-Ospelt G, Carlomagno T (2015) RNA structure determination by solid-state NMR spectroscopy. Nat Commun. doi:10.1038/Ncomms8024
Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15:8783–8798
Mori S, Abeygunawardana C, Johnson MO, van Zijl PC (1995) Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B 108:94–98
Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR (2002) Genetic control by a metabolite binding mRNA. Chem Biol 9:1043
Neuner S, Santner T, Kreutz C, Micura R (2015) The “speedy” synthesis of atom-specific N-15 imino/amido-labeled RNA. Chem-Eur J 21:11634–11643. doi:10.1002/chem.201501275
Nikonowicz EP, Sirr A, Legault P, Jucker FM, Baer LM, Pardi A (1992) Preparation of C-13 and N-15 Labeled Rnas for Heteronuclear Multidimensional Nmr-Studies. Nucleic Acids Res 20:4507–4513. doi:10.1093/nar/20.17.4507
Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA (2001) RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA 98:4899–4903. doi:10.1073/pnas.081082398
Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J (2005) An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc Natl Acad Sci USA 102:1372–1377. doi:10.1073/pnas.0406347102
Olsen GL, Edwards TE, Deka P, Varani G, Sigurdsson ST, Drobny GP (2005) Monitoring tat peptide binding to TAR RNA by solid-state 31P-19F REDOR NMR. Nucleic Acids Res 33:3447–3454. doi:10.1093/nar/gki626
Piotto M, Saudek V, Sklenar V (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR 2:661–665
Puffer B, Kreutz C, Rieder U, Ebert MO, Konrat R, Micura R (2009) 5-Fluoro pyrimidines: labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy. Nucleic Acids Res 37:7728–7740. doi:10.1093/nar/gkp862
Quant S et al (1994) Chemical synthesis of C-13-labeled monomers for the solid-phase and template controlled enzymatic-synthesis of DNA and RNA oligomers. Tetrahedron Lett 35:6649–6652. doi:10.1016/S0040-4039(00)73458-7
Reif B et al (1997) Structural comparison of oligoribonucleotides and their 2′-deoxy-2′-fluoro analogs by heteronuclear NMR spectroscopy. Helv Chim Acta 80:1952–1971. doi:10.1002/hlca.19970800614
Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H (2010) Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res 38:3834–3847. doi:10.1093/nar/gkq124
Rinnenthal J, Buck J, Ferner J, Wacker A, Furtig B, Schwalbe H (2011) Mapping the landscape of RNA dynamics with NMR spectroscopy. Acc Chem Res 44:1292–1301. doi:10.1021/ar200137d
Sashital DG, Cornilescu G, Butcher SE (2004) U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol 11:1237–1242. doi:10.1038/Nsmb863
Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J (2007) Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew Chem Int Ed Engl 46:1212–1219. doi:10.1002/anie.200604163
Scott LG, Geierstanger BH, Williamson JR, Hennig M (2004) Enzymatic synthesis and F-19 NMR studies of 2-fluoroadenine-substituted RNA. J Am Chem Soc 126:11776–11777. doi:10.1021/Ja047556x
Serganov A et al (2004) Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol 11:1729–1741. doi:10.1016/j.chembiol.2004.11.018
Shaka AJ, Barker PB, Freeman R (1985) Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson 64:547–552. doi:10.1016/0022-2364(85)90122-2
Sklenar V, Bax A (1987) Spin-echo water suppression for the generation of pure-phase two-dimensional NMR-spectra. J Magn Reson 74:469–479. doi:10.1016/0022-2364(87)90269-1
Sklenar V, Piotto M, Leppik R, Saudek V (1993) Gradient-tailored water suppression for H-1-N-15 Hsqc experiments optimized to retain full sensitivity. J Magn Reson Ser A 102:241–245. doi:10.1006/jmra.1993.1098
Staple DW, Butcher SE (2003) Solution structure of the HIV-1 frameshift inducing stem-loop RNA. Nucleic Acids Res 31:4326–4331. doi:10.1093/Nar/Gkg654
Suck D, Saenger W, Main P, Germain G, Declercq JP (1974) X-ray structure of 3′,5′-diacetyl-2′-deoxy-2′-fluorouridine: a pyrimidine nucleoside in the syn conformation. Biochim Biophys Acta 361:257–265
Thakur CS, Dayie TK (2012) Asymmetry of C-13 labeled 3-pyruvate affords improved site specific labeling of RNA for NMR spectroscopy. J Biomol NMR 52:65–77. doi:10.1007/s10858-011-9582-5
Wacker A, Buck J, Mathieu D, Richter C, Wohnert J, Schwalbe H (2011) Structure and dynamics of the deoxyguanosine-sensing riboswitch studied by NMR-spectroscopy. Nucleic Acids Res 39:6802–6812. doi:10.1093/nar/gkr238
Wacker A, Buck J, Richter C, Schwalbe H, Wohnert J (2012) Mechanisms for differentiation between cognate and near-cognate ligands by purine riboswitches. RNA Biol 9:672–680. doi:10.4161/rna.20106
Wenter P, Reymond L, Auweter SD, Allain FHT, Pitsch S (2006) Short, synthetic and selectively C-13-labeled RNA sequences for the NMR structure determination of protein-RNA complexes. Nucleic Acids Res. doi:10.1093/nar/gkl427
Yu C, Levy GC (1984) Two-dimensional heteronuclear NOE (HOESY) experiments: investigation of dipolar interactions between heteronuclei and nearby protons. J Am Chem Soc 106:6533–6537. doi:10.1021/Ja00334a013
Zhao C, Devany M, Greenbaum NL (2014) Measurement of chemical exchange between RNA conformers by (19)F NMR. Biochem Biophys Res Commun 453:692–695. doi:10.1016/j.bbrc.2014.09.075
Acknowledgments
We thank E. Stirnal for HPLC purification of RNAs. The work was supported within the DFG-funded collaborative research center: SFB902. H.S. is member of the DFG-funded cluster of excellence: macromolecular complexes. BMRZ is supported by the state of Hesse.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sochor, F., Silvers, R., Müller, D. et al. 19F-labeling of the adenine H2-site to study large RNAs by NMR spectroscopy. J Biomol NMR 64, 63–74 (2016). https://doi.org/10.1007/s10858-015-0006-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-015-0006-9