Abstract
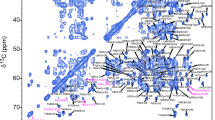
The icosahedral bacteriophage T7 is a 50 MDa double-stranded DNA (dsDNA) virus that infects Escherichia coli. Although there is substantial information on the physical and morphological properties of T7, structural information, based mostly on Raman spectroscopy and cryo-electron microscopy, is limited. Here, we apply the magic-angle spinning (MAS) solid-state NMR (SSNMR) technique to study a uniformly 13C and 15N labeled wild-type T7 phage. We describe the details of the large-scale preparation and purification of an isotopically enriched phage sample under fully hydrated conditions, and show a complete 13C and a near-complete 15N nucleotide-type specific assignment of the sugar and base moieties in the 40 kbp dsDNA of T7 using two-dimensional 13C–13C and 15N–13C correlation experiments. The chemical shifts are interpreted as reporters of a B-form conformation of the encapsulated dsDNA. While MAS SSNMR was found to be extremely useful in determining the structures of proteins in native-like environments, its application to nucleic acids has lagged behind, leaving a missing 13C and 15N chemical shift database. This work therefore expands the 13C and 15N database of real B-form DNA systems, and opens routes to characterize more complex nucleic acid systems by SSNMR.





Similar content being viewed by others
References
Abramov G, Morag O, Goldbourt A (2011) Magic-angle spinning NMR of a class I filamentous bacteriophage virus. J Phys Chem B 115:9671–9680
Aeschbacher T, Schmidt E, Blatter M et al (2013) Automated and assisted RNA resonance assignment using NMR chemical shift statistics. Nucleic Acids Res 41:e172
Agirrezabala X, Martín-Benito J, Castón JR et al (2005) Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J 24:3820–3829
Aitken A, Learmonth M (1996) Protein determination by UV absorption. In: Walker JM (ed) Protein Protoc. Handb. Humana Press, Totowa, pp 3–6
Asami S, Rakwalska-Bange M, Carlomagno T, Reif B (2013) Protein-RNA interfaces probed by 1H-detected MAS solid-state NMR spectroscopy. Angew Chem Int Ed Engl 52:2345–2349
Barton S, Heng X, Johnson BA, Summers MF (2013) Database proton NMR chemical shifts for RNA signal assignment and validation. J Biomol NMR 55:33–46
Ben-Shaul A (2013) Entropy, energy, and bending of DNA in viral capsids. Biophys J 104:L15–L17
Berjanskii MV, Neal S, Wishart DS (2006) PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res 34:W63–W69
Black LW, Thomas JA (2012) Condensed genome structure. Adv Exp Med Biol 726:469–487
Borer PN, LaPlante SR, Zanatta N, Levy GC (1988) Hydrogen-bonding effects and 13C-NMR of the DNA double helix. Nucleic Acids Res 16:2323–2332
Cady SD, Schmidt-Rohr K, Wang J et al (2010) Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 463:689–692
Cerritelli M, Cheng N, Rosenberg A et al (1997) Encapsidated conformation of bacteriophage T7 DNA. Cell 91:271–280
Cerritelli ME, Trus BL, Smith CS et al (2003) A second symmetry mismatch at the portal vertex of bacteriophage T7: 8-fold symmetry in the procapsid core. J Mol Biol 327:1–6
Cherepanov AV, Glaubitz C, Schwalbe H (2010) High-resolution studies of uniformly 13C,15N-labeled RNA by solid-state NMR spectroscopy. Angew Chem Int Ed Engl 49:4747–4750
Cho BP, Evans FE (1991) Structure of oxidatively damaged nucleic acid adducts. 3. Tautomerism, ionization and protonation of 8-hydroxyadenosine studied by 15N NMR spectroscopy. Nucleic Acids Res 19:1041–1047
Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13:289–302
Cuervo A, Pulido-Cid M, Chagoyen M et al (2013) Structural characterization of the bacteriophage T7 tail machinery. J Biol Chem 288:26290–26299
Daudén MI, Martín-Benito J, Sánchez-Ferrero JC et al (2013) Large terminase conformational change induced by connector binding in bacteriophage T7. J Biol Chem 288:16998–17007
Day LA, Marzec CJ, Reisberg SA, Casadevall A (1988) DNA packing in filamentous bacteriophages. Annu Rev Biophys Biophys Chem 17:509–539
Dayie KT (2005) Resolution enhanced homonuclear carbon decoupled triple resonance experiments for unambiguous RNA structural characterization. J Biomol NMR 32:129–139
De Groot HJ (2000) Solid-state NMR spectroscopy applied to membrane proteins. Curr Opin Struct Biol 10:593–600
Delaglio F, Grzesiek S, Vuister GW et al (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Deng H, Bloomfield V, Benevides J, Thomas G (1999) Dependence of the Raman signature of genomic B-DNA on nucleotide base sequence. Biopolymers 50:656–666
Dieckmann T, Feigon J (1997) Assignment methodology for larger RNA oligonucleotides: application to an ATP-binding RNA aptamer. J Biomol NMR 9:259–272
Duda RL, Ross PD, Cheng N et al (2009) Structure and energetics of encapsidated DNA in bacteriophage HK97 studied by scanning calorimetry and cryo-electron microscopy. J Mol Biol 391:471–483
Ebrahimi M, Rossi P, Rogers C, Harbison GS (2001) Dependence of 13C NMR chemical shifts on conformations of RNA nucleosides and nucleotides. J Magn Reson 150:1–9
Farès C, Amata I, Carlomagno T (2007) 13C-detection in RNA bases: revealing structure-chemical shift relationships. J Am Chem Soc 129:15814–15823
Fernández C, Szyperski T, Ono A et al (1998) NMR with 13C, 15N-doubly-labeled DNA: the Antennapedia homeodomain complex with a 14-mer DNA duplex. J Biomol NMR 12:25–37
Fiala R, Munzarová M, Sklenář V (2004) Experiments for correlating quaternary carbons in RNA bases. J Biomol NMR 29:477–490
Fonville JM, Swart M, Vokáčová Z et al (2012) Chemical shifts in nucleic acids studied by density functional theory calculations and comparison with experiment. Chem Eur J 18:12372–12387
Fürtig B, Richter C, Wöhnert J, Schwalbe H (2003) NMR spectroscopy of RNA. ChemBioChem 4:936–962
Fürtig B, Richter C, Bermel W, Schwalbe H (2004) New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J Biomol NMR 28:69–79
Ghose R, Marino JP, Wiberg KB, Prestegard JH (1994) Dependence of 13C chemical shifts on glycosidic torsional angles in ribonucleic acids. J Am Chem Soc 116:8827–8828
Goddard TD, Kneller DG (2008) SPARKY 3. University of California, San Francisco
Goldbourt A, Gross BJ, Day LA, McDermott AE (2007) Filamentous phage studied by magic-angle spinning NMR: resonance assignment and secondary structure of the coat protein in Pf1. J Am Chem Soc 129:2338–2344
Goldfarb A, Saidel L, Mosovich E (1951) The ultraviolet absorption spectra of proteins. J Biol Chem 193:397–404
Greene KL, Wang Y, Live D (1995) Influence of the glycosidic torsion angle on 13C and 15N shifts in guanosine nucleotides: investigations of G-tetrad models with alternating syn and anti bases. J Biomol NMR 5:333–338
Han Y, Hou G, Suiter CL et al (2013) Magic angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV-1 capsid protein assemblies. J Am Chem Soc 135:17793–17803
Henderson JT, Benight AS, Hanlon S (1992) A semi-micromethod for the determination of the extinction coefficients of duplex and single-stranded DNA. Anal Biochem 201:17–29
Hing AW, Vega S, Schaefer J (1992) Transferred-echo double-resonance NMR. J Magn Reson 96:205–209
Hu JZ, Facelli JC, Alderman DW et al (1998) 15N Chemical shift tensors in nucleic acid bases. J Am Chem Soc 120:9863–9869
Ionel A, Velázquez-Muriel JA, Luque D et al (2011) Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7. J Biol Chem 286:234–242
Jaroniec CP, MacPhee CE, Bajaj VS et al (2004) High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci U S A 101:711–716
Jehle S, Falb M, Kirkpatrick JP et al (2010) Intermolecular protein-RNA interactions revealed by 2D 31P–15N magic angle spinning solid-state NMR spectroscopy. J Am Chem Soc 132:3842–3846
Jiang W, Chang J, Jakana J et al (2006) Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612–616
Lam SL, Chi LM (2010) Use of chemical shifts for structural studies of nucleic acids. Prog Nucl Magn Reson Spectrosc 56:289–310
LaPlante SR, Boudreau EA, Zanatta N et al (1988) 13C NMR of the bases of three DNA oligonucleotide duplexes: assignment methods and structural features. Biochemistry 27:7902–7909
Leppert J, Urbinati CR, Häfner S et al (2004) Identification of NH…N hydrogen bonds by magic angle spinning solid state NMR in a double-stranded RNA associated with myotonic dystrophy. Nucleic Acids Res 32:1177–1183
Liu A, Majumdar A, Hu W et al (2000) NMR detection of NH···OC hydrogen bonds in 13C, 15N-labeled nucleic acids. J Am Chem Soc 122:3206–3210
Lopez del Amo JM, Schmidt M, Fink U et al (2012) An asymmetric dimer as the basic subunit in Alzheimer’s disease amyloid β fibrils. Angew Chem Int Ed Engl 51:6136–6139
Loquet A, Sgourakis NG, Gupta R et al (2012) Atomic model of the type III secretion system needle. Nature 486:276–279
Malináková K, Novosadová L, Lahtinen M et al (2010) 13C chemical shift tensors in hypoxanthine and 6-mercaptopurine: effects of substitution, tautomerism, and intermolecular interactions. J Phys Chem A 114:1985–1995
Marchanka A, Simon B, Carlomagno T (2013) A suite of solid-state NMR experiments for RNA intranucleotide resonance assignment in a 21 kDa protein-RNA complex. Angew Chemie 125:10180–10185
McDermott A, Gu Z (1996) Carbon and nitrogen chemical shifts: applications to solid state proteins. Encycl Nucl Magn Reson 1137–1147
Morag O, Abramov G, Goldbourt A (2014) Complete chemical shift assignment of the ssDNA in the filamentous bacteriophage fd reports on its conformation and on its interface with the capsid shell. J Am Chem Soc 136:2292–2301
Morcombe CR, Zilm KW (2003) Chemical shift referencing in MAS solid state NMR. J Magn Reson 162:479–486
Morcombe CR, Gaponenko V, Byrd RA, Zilm KW (2004) Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc 126:7196–7197
Ohlenschläger O, Haumann S, Ramachandran R, Görlach M (2008) Conformational signatures of 13C chemical shifts in RNA ribose. J Biomol NMR 42:139–142
Overman SA, Aubrey KL, Reilly KE et al (1998) Conformation and interactions of the packaged double-stranded DNA genome of bacteriophage T7. Biospectroscopy 4:S47–S56
Padrta P, Stefl R, Králík L et al (2002) Refinement of d(GCGAAGC) hairpin structure using one- and two-bond residual dipolar couplings. J Biomol NMR 24:1–14
Park SH, Das BB, Casagrande F et al (2012) Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 491:779–783
Pervushin K, Ono A, Fernández C et al (1998) NMR scalar couplings across Watson-Crick base pair hydrogen bonds in DNA observed by transverse relaxation-optimized spectroscopy. Proc Natl Acad Sci U S A 95:14147–14151
Petkova AT, Ishii Y, Balbach JJ et al (2002) A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A 99:16742–16747
Petkova AT, Yau W-M, Tycko R (2006) Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry 45:498–512
Petrov AS, Boz MB, Harvey SC (2007) The conformation of double-stranded DNA inside bacteriophages depends on capsid size and shape. J Struct Biol 160:241–248
Pines A, Gibby M, Waugh J (1973) Proton-enhanced NMR of dilute spins in solids. J Chem Phys 59:569–590
Porterfield JZ, Zlotnick A (2010) A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology 407:281–288
Richter C, Kovacs H, Buck J et al (2010) 13C-direct detected NMR experiments for the sequential J-based resonance assignment of RNA oligonucleotides. J Biomol NMR 47:259–269
Riedel K, Leppert J, Häfner S et al (2004) Homonuclear chemical shift correlation in rotating solids via RN νN symmetry-based adiabatic RF pulse schemes. J Biomol NMR 30:389–395
Riedel K, Leppert J, Ohlenschläger O et al (2005) TEDOR with adiabatic inversion pulses: resonance assignments of 13C/15N labelled RNAs. J Biomol NMR 31:49–57
Rossi P, Harbison GS (2001) Calculation of 13C chemical shifts in RNA nucleosides: structure-13C chemical shift relationships. J Magn Reson 151:1–8
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Santos RA, Tang P, Harbison GS (1989) Determination of the DNA sugar pucker using 13C NMR spectroscopy. Biochemistry 28:9372–9378
Schmid F (2001) Biological Macromolecules: UV-visible Spectrophotometry. Encycl Life Sci 1–4
Sergeyev IV, Day LA, Goldbourt A, McDermott AE (2011) Chemical shifts for the unusual DNA structure in Pf1 bacteriophage from dynamic-nuclear-polarization-enhanced solid-state NMR spectroscopy. J Am Chem Soc 133:20208–20217
Shen Y, Vernon R, Baker D, Bax A (2009) De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR 43:63–78
Stejskal EO, Schaefer J, Waugh J (1977) Magic-angle spinning and polarization transfer in proton-enhanced NMR. J Magn Reson 28:105–112
Studier FW (1969) The genetics and physiology of bacteriophage T7. Virology 39:562–574
Takegoshi K, Nakamura S, Terao T (2001) 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett 344:631–637
Tang M, Comellas G, Rienstra CM (2013) Advanced solid-state NMR approaches for structure determination of membrane proteins and amyloid fibrils. Acc Chem Res 46:2080–2088
Tataurov AV, You Y, Owczarzy R (2008) Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys Chem 133:66–70
Thakur RS, Kurur ND, Madhu PK (2006) Swept-frequency two-pulse phase modulation for heteronuclear dipolar decoupling in solid-state NMR. Chem Phys Lett 426:459–463
Thomas GJ, Serwer P (1990) Secondary structure of the double-stranded DNA genome of bacteriophage T7 in packaged, underpackaged and unpackaged states. J Raman Spectrosc 21:569–575
Tomar S, Green MM, Day LA (2007) DNA-protein interactions as the source of large-length-scale chirality evident in the liquid crystal behavior of filamentous bacteriophages. J Am Chem Soc 129:3367–3375
Tóth K, Bolard J, Rontó G, Aslanian D (1984) UV-induced small structural changes in the T7 bacteriophage studied by melting methods. Biophys Struct Mech 10:229–239
Ulrich EL, Akutsu H, Doreleijers JF et al (2008) BioMagResBank. Nucleic Acids Res 36:D402–D408
Veshtort M, Griffin RG (2006) SPINEVOLUTION: a powerful tool for the simulation of solid and liquid state NMR experiments. J Magn Reson 178:248–282
Wang S, Munro RA, Shi L et al (2013) Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat Methods 10:1007–1012
Wasmer C, Lange A, Van Melckebeke H et al (2008) Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science 319:1523–1526
Wijmenga SS, van Buuren BNM (1998) The use of NMR methods for conformational studies of nucleic acids. Prog Nucl Magn Reson Spectrosc 32:287–387
Wishart DS, Sykes BD (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4:171–180
Xu X-P, Chiu W-LAK, Au-Yeung SCF (1998) Chemical shift and structure relationship in nucleic acids: correlation of backbone torsion angles γ and α with 13C chemical shifts. J Am Chem Soc 120:4230–4231
Yu T-Y, Schaefer J (2008) REDOR NMR characterization of DNA packaging in bacteriophage T4. J Mol Biol 382:1031–1042
Acknowledgments
We would like to thank Dr. Udi Qimron from the Department of Human Microbiology, Faculty of Medicine in Tel Aviv University, for providing initial stocks of wild-type T7 phage and its host. We thank Anat Haimovich for the TEDOR simulations. Financial support was provided by the Israel Science Foundation. Partial support for the spectrometer was given by the center for Nanoscience and Nanotechnology of Tel Aviv University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abramov, G., Goldbourt, A. Nucleotide-type chemical shift assignment of the encapsulated 40 kbp dsDNA in intact bacteriophage T7 by MAS solid-state NMR. J Biomol NMR 59, 219–230 (2014). https://doi.org/10.1007/s10858-014-9840-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-014-9840-4