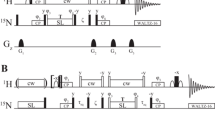
Abstract
The DREAM scheme is an efficient adiabatic homonuclear polarization-transfer method suitable for multi-dimensional experiments in biomolecular solid-state NMR. The bandwidth and dynamics of the polarization transfer in the DREAM experiment depend on a number of experimental and spin-system parameters. In order to obtain optimal results, the dependence of the cross-peak intensity on these parameters needs to be understood and carefully controlled. We introduce a simplified model to semi-quantitatively describe the polarization-transfer patterns for the relevant spin systems. Numerical simulations for all natural amino acids (except tryptophane) show the dependence of the cross-peak intensities as a function of the radio-frequency-carrier position. This dependency can be used as a guide to select the desired conditions in protein spectroscopy. Practical guidelines are given on how to set up a DREAM experiment for optimized Cα/Cβ transfer, which is important in sequential assignment experiments.







Similar content being viewed by others
Abbreviations
- NMR:
-
Nuclear magnetic resonance
- MAS:
-
Magic-angle spinning
- CP:
-
Cross-polarization
- DREAM:
-
Dipolar recoupling enhanced by amplitude modulation
References
Allen FH (2002) The Cambridge structural database: a quarter of a million crystal structures and rising. Acta Crystallogr B 58(Pt 3 Pt 1):380–388
Cheng VB, Suzukawa HH, Wolfsberg M (1973) Investigations of a nonrandom numerical-method for multidimensional integration. J Chem Phys 59(8):3992–3999
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMR-Pipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Detken A, Hardy E, Ernst M, Meier BH (2002) Simple and efficient decoupling in magic-angle spinning solid- state NMR: the XiX scheme. Chem Phys Lett 356(3–4):298–304
Ernst M, Meier BH (2010) Adiabatic polarization-transfer methods in MAS spectroscopy. Encycl Magn Reson (EMR) online: doi:10.1002/9780470034590.emrstm9780470030004.pub9780470034592
Fung B, Khitrin A, Ermolaev K (2000) An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson 142(1):97–101
Gath J, Habenstein B, Bousset L, Melki R, Meier BH, Böckmann A (2011) Solid-state NMR sequential assignments of α-synuclein. Biomol NMR Assign: (in press, available on the web)
Habenstein B, Wasmer C, Bousset L, Sourigues Y, Schütz A, Loquet A, Meier BH, Melki R, Böckmann A (2011) Extensive de novo solid-state NMR assignments of the 33 kDa C-terminal domain of the Ure2 prion. J Biomol NMR 51(3):235–243
Hediger S, Meier BH, KURUR N, BODENHAUSEN G, Ernst R (1994) NMR cross polarization by adiabatic passage through the Hartmann-Hahn condition (APHH). Chem Phys Lett 223:283–288
Hediger S, Meier BH, Ernst R (1995) Adiabatic passage Hartmann-Hahn cross polarization in NMR under magic angle sample spinning. Chem Phys Lett 240:449
Jeener J, Meier BH, Bachmann P, Ernst RR (1979) Investigation of exchange processes by two-dimensional NMR spectroscopy. J Chem Phys 71(11):4546
Kneller D, Kuntz I (1993) UCSF sparky—an NMR display, annotation and assignment tool. J Cell Biochem 53:254
Loquet A, Bousset L, Gardiennet C, Sourigues Y, Wasmer C, Habenstein B, Schütz A, Meier BH, Melki R, Böckmann A (2009) Prion fibrils of Ure2p assembled under physiological conditions contain highly ordered, natively folded modules. J Mol Biol 394(1):108–118
Marion D, Wüthrich K (1983) Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun 113:967–974
Radloff C, Ernst R (1989) Spin topology filtration in NMR. Mol Phys 66(1):161–197
Schuetz A, Wasmer C, Habenstein B, Verel R, Greenwald J, Riek R, Böckmann A, Meier BH (2010) Protocols for the sequential solid-state NMR spectroscopic assignment of a uniformly labeled 25 kDa protein: HET-s(1-227). ChemBioChem 11(11):1543–1551
Shi L, Kawamura I, Jung K-H, Brown LS, Ladizhansky V (2010) Conformation of a seven-helical transmembrane photosensor in the lipid environment. Angew Chem Int Ed Engl 50(6):1302–1305
Smith S, Levante T, Meier BH, Ernst R (1994) Computer simulations in magnetic resonance: an object oriented programming approach. J Magn Reson Ser A 106:75–105
Sperling LJ, Berthold DA, Sasser TL, Jeisy-Scott V, Rienstra CM (2010) Assignment strategies for large proteins by magic-angle spinning NMR: the 21-kDa disulfide-bond-forming enzyme DsbA. J Mol Biol 399(2):268–282
Verel R (2001) Adiabatic methods for homonuclear dipolar recoupling in magic angle spinning solid-state NMR. DISS ETH 14152
Verel R, Meier BH (2004) Polarization-transfer methods in solid-state magic-angle- spinning NMR: adiabatic CN pulse sequences. ChemPhysChem 5(6):851–862
Verel R, Baldus M, Ernst M, Meier BH (1998) A homonuclear spin-pair filter for solid-state NMR based on adiabatic-passage techniques. Chem Phys Lett 287:421–428
Verel R, Ernst M, Meier BH (2001) Adiabatic dipolar recoupling in solid-state NMR: the DREAM scheme. J Magn Reson 150(1):81–99
Wang Y, Jardetzky O (2002) Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci 11(4):852–861
Wasmer C, Schütz A, Loquet A, Buhtz C, Greenwald J, Riek R, Böckmann A, Meier BH (2009) The molecular organization of the fungal prion HET-s in its amyloid form. J Mol Biol 394(1):119–127
Westfeld T (2010) Polarization transfer dynamics in multi-spin systems using the DREAM scheme. Diss ETH 19196:1–153
Ye C, Fu R, Hu J, HOU L, Ding S (1993) C-13 chemical-shift anisotropies of solid amino-acids. Magn Reson Chem 31(8):699–704
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Westfeld, T., Verel, R., Ernst, M. et al. Properties of the DREAM scheme and its optimization for application to proteins. J Biomol NMR 53, 103–112 (2012). https://doi.org/10.1007/s10858-012-9627-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-012-9627-4