Abstract
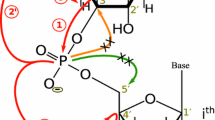
A five-dimensional (5D) APSY (automated projection spectroscopy) HCNCH experiment is presented, which allows unambiguous correlation of sugar to base nuclei in nucleic acids. The pulse sequence uses multiple quantum (MQ) evolution which enables long constant-time evolution periods in all dimensions, an improvement that can also benefit non-APSY applications. Applied with an RNA with 23 nucleotides the 5D APSY-HCNCH experiment produced a complete and highly precise 5D chemical shift list within 1.5 h. Alternatively, and for molecules where the out-and-stay 5D experiment sensitivity is not sufficient, a set of out-and-back 3D APSY-HCN experiments is proposed: an intra-base (3D APSY-b-HCN) experiment in an MQ or in a TROSY version, and an MQ sugar-to-base (3D APSY-s-HCN) experiment. The two 3D peak lists require subsequent matching via the N1/9 chemical shift values to one 5D peak list. Optimization of the 3D APSY experiments for maximal precision in the N1/9 dimension allowed matching of all 15N chemical shift values contained in both 3D peak lists. The precise 5D chemical shift correlation lists resulting from the 5D experiment or a pair of 3D experiments also provide a valuable basis for subsequent connection to chemical shifts derived with other experiments.




Similar content being viewed by others
References
Allain FHT, Duss O, Maris C, von Schroetter C (2010) A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic Acids Res 38(20). doi:10.1093/nar/gkq756
Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR (1992) Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res 20(17):4515–4523
Bax A, Freeman R (1981) Investigation of complex networks of spin–spin coupling by two-dimensional NMR. J Magn Reson 44(3):542–561
Brutscher B, Simorre JP (2001) Transverse relaxation optimized HCN experiment for nucleic acids: combining the advantages of TROSY and MQ spin evolution. J Biomol NMR 21(4):367–372
Carlomagno T, Fares C (2006) SHARP-TACSY: triple-band tailored correlated spectroscopy for base-to-sugar transfer in nucleic acid residues with intermediate time scale motions. J Am Chem Soc 128(30):9856–9862
Emsley L, Bodenhausen G (1990) Gaussian pulse cascades—new analytical functions for rectangular selective inversion and in-phase excitation in NMR. Chem Phys Lett 165(6):469–476
Farmer BT, Müller L, Nikonowicz EP, Pardi A (1993) Unambiguous resonance assignments in 13C, 15N-labeled nucleic acids by 3D triple resonance NMR. J Am Chem Soc 115(23):11040–11041
Farmer BT, Müller L, Nikonowicz EP, Pardi A (1994) Unambiguous through-bond sugar-to-base correlations for purines in 13C, 15N-labeled nucleic acids—the HsCsNb, HsCs(N)bCb, and HbNbCb experiments. J Biomol NMR 4(1):129–133
Fiala R, Jiang F, Sklenai V (1998) Sensitivity optimized HCN and HCNCH experiments for 13C/15N labeled oligonucleotides. J Biomol NMR 12(3):373–383
Fiala R, Czernek J, Sklenar V (2000) Transverse relaxation optimized triple-resonance NMR experiments for nucleic acids. J Biomol NMR 16(4):291–302
Fiorito F, Hiller S, Wider G, Wüthrich K (2006) Automated resonance assignment of proteins: 6D APSY-NMR. J Biomol NMR 35(1):27–37
Flinders J, Dieckmann T (2006) NMR spectroscopy of ribonucleic acids. Prog Nucl Mag Res Sp 48(2–3):137–159
Fürtig B, Richter C, Wöhnert J, Schwalbe H (2003) NMR spectroscopy of RNA. Chembiochem 4(10):936–962
Fürtig B, Richter C, Bermel W, Schwalbe H (2004) New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J Biomol NMR 28(1):69–79
Geen H, Freeman R (1991) Band-selective radiofrequency pulses. J Magn Reson 93(1):93–141
Gossert AD, Hiller S, Fernandez C (2011) Automated NMR resonance assignment of large proteins for protein-ligand interaction studies. J Am Chem Soc 133(2):210–213
Guillerez J, Lopez PJ, Proux F, Launay H, Dreyfus M (2005) A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc Natl Acad Sci USA 102(17):5958–5963
Güntert P, Dötsch V, Wider G, Wüthrich K (1992) Processing of multidimensional NMR data with the new software PROSA. J Biomol NMR 2(6):619–629
Hiller S, Fiorito F, Wüthrich K, Wider G (2005) Automated projection spectroscopy (APSY). Proc Natl Acad Sci USA 102(31):10876–10881
Hiller S, Joss R, Wider G (2008a) Automated NMR assignment of protein side chain resonances using automated projection spectroscopy (APSY). J Am Chem Soc 130(36):12073–12079
Hiller S, Wider G, Wüthrich K (2008b) APSY-NMR with proteins: practical aspects and backbone assignment. J Biomol NMR 42(3):179–195
Hu WD, Gosser YQ, Xu WJ, Patel DJ (2001) Novel 2D and 3D multiple-quantum bi-directional HCNCH experiments for the correlation of ribose and base protons/carbons in 13C/15N labeled RNA. J Biomol NMR 20(2):167–172
Jaroniec CP, Boisbouvier J, Tworowska I, Nikonowicz EP, Bax A (2005) Accurate measurement of 15N–13C residual dipolar couplings in nucleic acids. J Biomol NMR 31(3):231–241
Kim S, Szyperski T (2003) GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc 125(5):1385–1393
Kupce E, Freeman R (2003) Projection-reconstruction of three-dimensional NMR spectra. J Am Chem Soc 125(46):13958–13959
Marino JP, Diener JL, Moore PB, Griesinger C (1997) Multiple-quantum coherence dramatically enhances the sensitivity of CH and CH2 correlations in uniformly 13C-labeled RNA. J Am Chem Soc 119(31):7361–7366
Marion D, Ikura M, Tschudin R, Bax A (1989) Rapid recording of 2D NMR spectra withoug phase cycling—application to the study of hydrogen-exchange in proteins. J Magn Reson 85(2):393–399
Narayanan RL, Dürr UHN, Bibow S, Biernat J, Mandelkow E, Zweckstetter M (2010) Automatic assignment of the intrinsically disordered protein Tau with 441-residues. J Am Chem Soc 132(34):11906–11907
Nyholm T, Andang M, Hotchkiss G, Hard T, Baumann H, Larsson S, Ahrlundrichter L (1995) A method for production of 13C/15N double-labeled RNA in Escherichia Coli, and subsequent in vitro synthesis of ribonucleotide 5′-triphosphates. J Biochem Biophys Methods 30(1):59–68
Pervushin KV, Wider G, Wüthrich K (1998) Single transition-to-single transition polarization transfer (ST2-PT) in 15N, 1H-TROSY. J Biomol NMR 12(2):345–348
Price SR, Oubridge C, Varani G, Nagai K (1998) Preparation of RNA:protein complexes for X-ray crystallography and NMR. In: Smith CWJ (ed) RNA-protein interactions: a practical approach, vol 192. Oxford University Press, Oxford, pp 37–74
Ravindranathan S, Kim CH, Bodenhausen G (2003) Cross correlations between 13C–1H dipolar interactions and 15N chemical shift anisotropy in nucleic acids. J Biomol NMR 27(4):365–375
Riek R, Pervushin K, Fernandez C, Kainosho M, Wüthrich K (2001) 13C, 13C- and 13C, 1H-TROSY in a triple resonance experiment for ribose-base and intrabase correlations in nucleic acids. J Am Chem Soc 123(4):658–664
Rinnenthal J, Richter C, Ferner J, Duchardt E, Schwalbe H (2007) Quantitative Gamma-HCNCH: determination of the glycosidic torsion angle chi in RNA oligonucleotides from the analysis of CH dipolar cross-correlated relaxation by solution NMR spectroscopy. J Biomol NMR 39(1):17–29
Shaka AJ, Keeler J (1987) Broadband spin decoupling in isotropic liquids. Prog Nucl Mag Res Sp 19:47–129
Sklenar V, Peterson RD, Rejante MR, Feigon J (1993a) 2D and 3D HCN experiments for correlating base and sugar resonances in 15N, 13C-labeled RNA oligonucleotides. J Biomol NMR 3(6):721–727
Sklenar V, Peterson RD, Rejante MR, Wang E, Feigon J (1993b) 2D triple-resonance HCNCH experiment for direct correlation of ribose H1′ and base H6/8 protons in 13C, 15N-labeled RNA oligonucleotides. J Am Chem Soc 115(25):12181–12182
Sklenar V, Dieckmann T, Butcher SE, Feigon J (1998) Optimization of triple-resonance HCN experiments for application to larger RNA oligonucleotides. J Magn Reson 130(1):119–124
Szyperski T, Wider G, Bushweller JH, Wüthrich K (1993) Reduced dimensionality in triple-resonance NMR experiments. J Am Chem Soc 115(20):9307–9308
Tate S, Ono A, Kainosho M (1994) An alternative triple-resonance method for the through-bond correlation of intranucleotide H1′ and H8 NMR signals of purine nucleotides. Application to a DNA dodecamer with fully 13C, 15N-labeled deoxyadenosine residues. J Am Chem Soc 116(13):5977–5978
Van Melckebeke H, Pardi A, Boisbouvier J, Simorre JP, Brutscher B (2005) Resolution-enhanced base-type-edited HCN experiment for RNA. J Biomol NMR 32(4):263–271
Wider G, Dreier L (2006) Measuring protein concentrations by NMR spectroscopy. J Am Chem Soc 128:2571–2576
Yan JL, Corpora T, Pradhan P, Bushweller JH (2002) MQ-HCN-based pulse sequences for the measurement of 13C1′–1H1′, 13C1′–15N, 1H1′–15N, 13C1′–13C2′, 1H1′–13C2′, 13C6/8–1H6/8, 13C6/8–15N, 1H6/8–15N, 13C6–13C5, 1H6–13C5 dipolar couplings in 13C, 15N-labeled DNA (and RNA). J Biomol NMR 22(1):9–20
Acknowledgments
We thank Prof. Sebastian Hiller (University Basel) for technical discussions, and Prof. Frédéric Allain (ETH Zurich) for the provision of [13C, 15N]-labeled nucleotides. The Swiss National Fund (SNF) is gratefully acknowledged for financial support (project 200021_120048).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krähenbühl, B., Hofmann, D., Maris, C. et al. Sugar-to-base correlation in nucleic acids with a 5D APSY-HCNCH or two 3D APSY-HCN experiments. J Biomol NMR 52, 141–150 (2012). https://doi.org/10.1007/s10858-011-9588-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-011-9588-z