Abstract
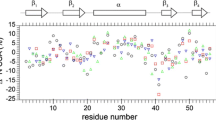
Residue-specific amide proton spin-flip rates K were measured for peptide-free and peptide-bound calmodulin. K approximates the sum of NOE build-up rates between the amide proton and all other protons. This work outlines the theory of multi-proton relaxation, cross relaxation and cross correlation, and how to approximate it with a simple model based on a variable number of equidistant protons. This model is used to extract the sums of K-rates from the experimental data. Error in K is estimated using bootstrap methodology. We define a parameter Q as the ratio of experimental K-rates to theoretical K-rates, where the theoretical K-rates are computed from atomic coordinates. Q is 1 in the case of no local motion, but decreases to values as low as 0.5 with increasing domination of sidechain protons of the same residue to the amide proton flips. This establishes Q as a monotonous measure of local dynamics of the proton network surrounding the amide protons. The method is applied to the study of proton dynamics in Ca2+-saturated calmodulin, both free in solution and bound to smMLCK peptide. The mean Q is 0.81 ± 0.02 for free calmodulin and 0.88 ± 0.02 for peptide-bound calmodulin. This novel methodology thus reveals the presence of significant interproton disorder in this protein, while the increase in Q indicates rigidification of the proton network upon peptide binding, confirming the known high entropic cost of this process.












Similar content being viewed by others
References
Akke M, Bruschweiler R, Palmer AG (1993) NMR order parameters and free-energy—an analytical approach and its application to cooperative Ca2 + binding by calbindin-D(9 k). J Am Chem Soc 115:9832–9833
Best RB, Clarke J, Karplus M (2005) What contributions to protein side-chain dynamics are probed by NMR experiments? A molecular dynamics simulation analysis. J Mol Biol 349:185–203
Brüschweiler R, Roux B, Blackledge M, Griesinger C, Karplus M, Ernst R (1992) Influence of rapid intramolecular motion on NMR cross-relaxation rates. A molecular dynamics study of antamanide in solution. J Am Chem Soc 114:2289–2302
Cooper A, Dryden D (1984) Allostery without conformational change. A plausible model. Eur Biophys J 11:103–109
Crivici A, Ikura M (1995) Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct 24:85–116
Cruickshank D (1999) Remarks about protein structure precision. Acta Cryst D55:583–601
Efron B, Tibshirani R (1994) An introduction to the bootstrap. Chapman & Hall/CRC, London
Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D (2005) Intrinsic dynamics of an enzyme underlies catalysis. Nature 438:117–121
Ferrage F, Pelupessy P, Cowburn D, Bodenhausen G (2006) Protein backbone dynamics through 13C’-13C alpha cross-relaxation in NMR spectroscopy. J Am Chem Soc 128:11072–11078
Fischer MWF, Zeng L, Pang Y, Hu W, Majumdar A, Zuiderweg ERP (1997) Experimental characterization of models for backbone pico-second dynamics in proteins. Quantification of NMR auto- and cross correlation relaxation mechanisms involving different nuclei of the peptide plane. J Am Chem Soc 119:12629–12642
Frederick KK, Marlow MS, Valentine KG, Wand AJ (2007) Conformational entropy in molecular recognition by proteins. Nature 448:325–329
Gong Q, Ishima R (2007) 15 N–1H NOE experiment at high magnetic field strengths. J Biomol NMR 37:147–157
Kroenke CD, Loria JP, Lee LK, Rance M, Palmer AGIII (1998) Longitudinal and transverse 1H–15 N dipolar/15 N chemical shift anisotropy relaxation interference: unambiguous determination of rotational diffusion tensors and chemical exchange effects in biological macromolecules. J Am Chem Soc 120:7905–7915
Labeikovsky W, Eisenmesser EZ, Bosco DA, Kern D (2007) Structure and dynamics of pin1 during catalysis by NMR. J Mol Biol 367:1370–1381
Lee LK, Rance M, Chazin WJ, Palmer AGIII (1997) Rotational diffusion anisotropy of proteins from simultaneous analysis of 15 N and 13C alpha nuclear spin relaxation. J Biomol NMR 9:287–298
Lee AL, Kinnear SA, Wand AJ (2000) Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nat Struct Biol 7:72–77
Lindemann F (1910) The calculation of molecular vibration frequencies. Physikalisches Zeitschrift 11:609–612
Loria JP, Rance M, Palmer AGIII (1999) A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc 121:2331–2332
Lovell S, Davis I, Arendall W, De Bakker P, Word J, Prisant M, Richardson J, Richardson D (2003) Structure validation by C-alpha geometry: phi, psi, and C-beta deviation. Proteins Struct Funct Genet 50:437–450
Mäler L, Blankenship J, Rance M, Chazin WJ (2000) Site-site communication in the EF-hand Ca2 + -binding protein calbindin D9 k. Nat Struct Biol 7:245–250
Millet O, Muhandiram DR, Skrynnikov NR, Kay LE (2002) Deuterium spin probes of side-chain dynamics in proteins. 1. Measurement of five relaxation rates per deuteron in (13)C-labeled and fractionally (2)H-enriched proteins in solution. J Am Chem Soc 124:6439–6448
Ming D, Bruschweiler R (2004) Prediction of methyl-side chain dynamics in proteins. J Biomol NMR 29:363–368
Mittermaier A, Kay LE (2006) New tools provide new insights in NMR studies of protein dynamics. Science 312:224–228
Neuhaus D, Williamson M (2000) The nuclear Overhauser effect in stereochemical and conformational analysis. Wiley, New York
Olejniczak E, Dobson C, Karplus M, Levy R (1984) Motional averaging of proton nuclear Overhauser effects in proteins. Predictions from a molecular dynamics simulation of lysozyme. J Am Chem Soc 106:1923–1930
Palmer III AG (2009) Inertia and diffusion tensors. http://biochemistryhscolumbiaedu/labs/palmer/software/diffusionhtml
Pang A, Buck M, Zuiderweg ERP (2002) Backbone dynamics of the ribonuclease binase active site area using multinuclear (15N and 13CO) NMR relaxation and computational molecular dynamics. Biochemistry 41:2655–2666
Pellecchia M, Pang Y, Wang L, Kurochkin AV, Kumar A, Zuiderweg ERP (1999) Quantitative measurement of cross-correlations between 15N and 13CO chemical shift anisotropy relaxation mechanisms by multiple quantum NMR. J Am Chem Soc 121:9165–9170
Pelupessy P, Espallargas GM, Bodenhausen G (2003) Symmetrical reconversion: measuring cross-correlation rates with enhanced accuracy. J Magn Reson 161:258–264
Peti W, Meiler J, Bruschweiler R, Griesinger C (2002) Model-free analysis of protein backbone motion from residual dipolar couplings. J Am Chem Soc 124:5822–5833
Post C (1992) Internal motional averaging and three-dimensional structure determination by nuclear magnetic resonance. J Mol Biol 224:1087–1101
Post CB, Dobson CM, Karplus M (1989) A molecular dynamics analysis of protein structural elements. Proteins 5:337–354
Spera S, Ikura M, Bax A (1991) Measurement of the exchange rates of rapidly exchanging amide protons: application to the study of calmodulin and its complex with a myosin light chain kinase fragment. J Biomol NMR 1:155–165
Tang C, Iwahara J, Clore GM (2006) Visualization of transient encounter complexes in protein-protein association. Nature 444:383–386
Tropp J (1980) Dipolar relaxation and nuclear Overhauser effects in nonrigid molecules: the effect of fluctuating internuclear distances. J Chem Phys 72:6035–6043
Tugarinov V, Sprangers R, Kay LE (2007) Probing side-chain dynamics in the proteasome by relaxation violated coherence transfer NMR spectroscopy. J Am Chem Soc 129:1743–1750
Valentine K, Ng H, Schneeweis J, Kranz J, Frederick K, Alber T, Wand A (2009) Ultrahigh resolution crystal structure of calmodulin-smooth muscle light kinase peptide complex. PDB (to be published)
Vogel HJ (1994) The Merck Frosst award lecture 1994. Calmodulin: a versatile calcium mediator protein. Biochem Cell Biol 72:357–376
Wand AJ, Bieber RJ, Urbauer JL, McEvoy RP, Gan Z (1995) Carbon relaxation in randomly fractionally 13C-enriched proteins. J Magn Reson B 108:173–175
Wang L, Pang Y, Holder T, Brender JR, Kurochkin AV, Zuiderweg ER (2001) Functional dynamics in the active site of the ribonuclease binase. Proc Natl Acad Sci USA 98:7684–7689
Wang C, Rance M, Palmer A (2003) Mapping chemical exchange in proteins with MW > 50 kD. J Am Chem Soc 125:8968–8969
Weaver DS, Zuiderweg ER (2008) Eta(z)/kappa: a transverse relaxation optimized spectroscopy NMR experiment measuring longitudinal relaxation interference. J Chem Phys 128:155103
Werbelow L, Grant D (1977) Intramolecular dipolar relaxation in multispin systems. Adv Magn Reson 9:189–299
Wilson M, Brunger A (2000) The 1.0 A crystal structure of Ca(2+)-bound calmodulin: an analysis of disorder and implications for functionally relevant plasticity. J Mol Biol 301:1237–1256
Yang D, Kay LE (1996) Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: application to protein folding. J Mol Biol 263:369–382
Yang D, Mittermaier A, Mok YK, Kay LE (1998) A study of protein side-chain dynamics from new 2H auto-correlation and 13C cross-correlation NMR experiments: application to the N-terminal SH3 domain from drk. J Mol Biol 276:939–954
Zhang F, Bruschweiler R (2002) Contact model for the prediction of NMR N-H order parameters in globular proteins. J Am Chem Soc 124:12654–12655
Zhou Y, Vitkup D, Karplus M (1999) Native proteins are surface-molten solids: application of the Lindemann criterion for the solid versus liquid state. J Mol Biol 285:1371–1375
Acknowledgments
This work was supported by NIH grant GM63027 and a University of Michigan Rackham Pre-doctoral award to D. S. W. We thank Dr. A. J. Wand for the samples of calmodulin.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Weaver, D.S., Zuiderweg, E.R.P. Protein proton–proton dynamics from amide proton spin flip rates. J Biomol NMR 45, 99–119 (2009). https://doi.org/10.1007/s10858-009-9351-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-009-9351-x