Abstract
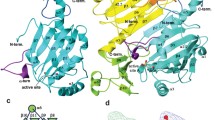
AF2241 is a hypothetical protein from Archaeoglobus fulgidus and it belongs to the PFam domain of unknown function 369 (DUF369). NMR structural determination reveals that AF2241 adopts a cyclophilin-like fold, with a β-barrel core composed of eight β-strands, one α-helix, and one 310 helix located at each end of the barrel. The protein displays a high structural similarity to TM1367, another member of DUF369 whose structure has been determined recently by X-ray crystallography. Structural similarity search shows that AF2241 also has a high similarity to human cyclophilin A, however, sequence alignment and electrostatic potential analysis reveal that the residues in the PPIase catalytic site of human cyclophilin A are not conserved in AF2241 or TM1367. Instead, a putative active site of AF2241 maps to a negatively charged pocket composed of 9 conserved residues. Our results suggest that although AF2241 adopts the same fold as the human cyclophilin A, it may have distinct biological function.


Similar content being viewed by others
References
Ai X, Semesi A, Yee A, Arrowsmith CH, Li SSC, Choy WY (2007) Backbone and side chain 1H, 13C, and 15N resonance assignments of AF2241 from Archaeoglobus fulgidus. J Biomol NMR 38:183
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32:D138–D141
Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst D54:905–921
Binkowski TA, Naghibzadeg S, Liang J (2003) CASTp: computed atlas of surface topography of proteins. Nucleic Acids Res 31:3352–3355
Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20:426–427
Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13:289–302
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Holm L, Sander C (1995) Dali – a network tool for protein-structure comparison Trends Biochem Sci 20:478–480
Huai Q, Kim HY, Liu Y, Zhao Y, Mondragon A, Liu JO, Ke H (2002) Crystal structure of calcineurin-cyclophlin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes. Proc Natl Acad Sci USA 99:12037–12042
Jin KK, Krishna SS, Schwarzenbacher R et al (2006) Crystal structure of TM1367 from Thermotoga maritima at 1.90 Å resolution reveals an atypical member of the cyclophilin (Peptidylprolyl Isomerase) fold. Proteins 63:1112–1118
Johnson BA, Blevins RA (1994) NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR 4:603–614
Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: A program for display and analysis of macromolecular structure. J Mol Graph 14:51–55
Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486
Livingstone CD, Barton GJ (1993) Protein sequence alignments: A strategy for the hierarchical analysis of residue conservation. CABIOS 9:745–756
Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247:536–540
Takahashi N, Hayano T, Suzuki M (1989) Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature 337:473–475
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B (2003) String: a database of predicted functional associations between proteins. Nucleic Acids Res 31:258–261
Wishart DS, Sykes BD (1994) The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4:171–180
Yanai I, DeLisi C (2002) The society of genes: networks of functional links between from comparative genomics. Genome Bio. 3, research0064.1–0064.12
Acknowledgements
We would like to thank Drs. Hsiu-Ju Tseng and Matthew Revington for helpful discussions. This work was supported by the Ontario Research Development Challenge Fund (to CHA and SSCL), Genome Canada, and a CIHR resource Grant.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ai, X., Li, L., Semesi, A. et al. Hypothetical protein AF2241 from Archaeoglobus fulgidus adopts a cyclophilin-like fold. J Biomol NMR 38, 353–358 (2007). https://doi.org/10.1007/s10858-007-9172-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-007-9172-8