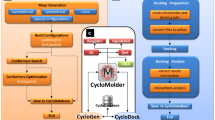
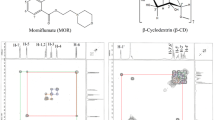
Abstract
In this review the authors present an overview of different molecular modeling campaigns dealing with the study and characterization of cyclodextrins (CDs) inclusion complexes with applicability to diverse biomedical and technological domains. The aim of this review is to present in a concise manner the new tendencies towards CDs molecular modeling studies in the context of a scientific computing era characterized by detailed and exhaustively validated molecular modeling protocols combined with and enormous and continuously growing computing power. Therefore, the present review covers research efforts reported in the last 5 years, including the simulation of native and modified CDs in a new and more detailed manner than what was possible in the past as well as their inclusion complexes with bioactive molecules studied by detailed protocols and exhaustive free-energy of binding calculations. Also, particular emphasis is devoted to the molecular modeling simulation of CDs included as part of drug delivery matrixes and intelligent nanodevices such as CD-based molecular motors.


Similar content being viewed by others
References
Crini, G.: Review: a history of cyclodextrins. Chem. Rev. 114(21), 10940–10975 (2014)
Sharma, N., Baldi, A.: Exploring versatile applications of cyclodextrins: an overview. Drug Deliv. 23(3), 729–747 (2016)
Iacovino, R., V Caso, J., Di Donato, C., Malgieri, G., Palmieri, M., Russo, L., Isernia, C.: Cyclodextrins as complexing agents: preparation and applications. Curr. Org. Chem. 21(2), 162–176 (2017)
Adeoye, O., Cabral-Marques, H.: Cyclodextrin nanosystems in oral drug delivery: a mini review. Int. J. Pharmaceut. 531(2), 521–531 (2017)
Barbour, L.: Experimental and computational methods in supramolecular chemistry. In: Gokel, G. W., Barbour, L. (eds.) Comprehensive Supramolecular Chemistry II, vol. 2. Elsevier, United Kingdom (2017)
Atwood, J.L., Lehn, J.M.: Comprehensive Supramolecular Chemistry: Cyclodextrins. Pergamon, Oxford (1996)
Jensen, J.H.: Molecular Modeling Basics. CRC Press, Boca Raton (2010)
Schlick, T.: Molecular Modeling and Simulation: An Interdisciplinary Guide. Springer, New York (2013)
Castro, E., Barbiric, D.: Molecular modeling and cyclodextrins: a relationship strengthened by complexes. Curr. Org. Chem. 10(7), 715–729 (2006)
Davies, J.: Spectroscopic and computational studies of supramolecular systems, vol. 4. Springer, London (2013)
Lipkowitz, K.B.: Applications of computational chemistry to the study of cyclodextrins. Chem. Rev. 98(5), 1829–1873 (1998)
Zhao, Q., Zhang, W., Wang, R., Wang, Y., Ouyang, D.: Research advances in molecular modeling in cyclodextrins. Curr. Pharm. Design. 23(3), 522–531 (2017)
Snir, M.: The future of supercomputing. In: Proceedings of the 2028th ACM International Conference on Supercomputing 14, pp. 261–262, ACM
Xie, X., Fang, X., Hu, S., Wu, D.: Evolution of supercomputers. Front. Comput. Sci. China 4(4), 428–436 (2010)
Maximova, T., Moffatt, R., Ma, B., Nussinov, R., Shehu, A.: Principles and overview of sampling methods for modeling macromolecular structure and dynamics. PLoS Comput. Biol. 12(4), e1004619 (2016)
Lameira, J., Kupchencko, I., Warshel, A.: Enhancing paradynamics for QM/MM sampling of enzymatic reactions. J. Phys. Chem. B 120(9), 2155–2164 (2016)
Cai, Y., See, S.: GPU Computing and Applications. Springer, Singapore (2015)
Ren, B., Zhang, M., Gao, H., Zheng, J., Jia, L.: Atomic elucidation of the cyclodextrin effects on DDT solubility and biodegradation. Phys. Chem. Chem. Phys. 18(26), 17380–17388 (2016)
Cerón-Carrasco, J.P., den-Haan, H., Peña-García, J., Contreras-García, J., Pérez-Sánchez, H.: Exploiting the cyclodextrins ability for antioxidants encapsulation: a computational approach to carnosol and carnosic acid embedding. Comput. Theor. Chem. 1077, 65–73 (2016)
Pagadala, N.S., Syed, K., Tuszynski, J.: Software for molecular docking: a review. Biophys. Rev. 9(2), 91–102 (2017)
Suárez, D., Díaz, N.: Conformational and entropy analyses of extended molecular dynamics simulations of α-, β-and γ-cyclodextrins and of the β-cyclodextrin/nabumetone complex. Phys. Chem. Chem. Phys. 19(2), 1431–1440 (2017)
Salomon-Ferrer, R., Götz, A.W., Poole, D., Le Grand, S., Walker, R.C.: Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J. Chem. Theory Comput. 9(9), 3878–3888 (2013)
Salomon-Ferrer, R., Case, D.A., Walker, R.C.: An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. 3(2), 198–210 (2013)
Ivanov, P.: Further studies on the conformations of large-ring cyclodextrins. Bulg. Chem. Commun. 46(A), 238–245 (2014)
Assaf, K.I., Gabel, D., Zimmermann, W., Nau, W.M.: High-affinity host-guest chemistry of large-ring cyclodextrins. Org. Biomol. Chem. 14(32), 7702–7706 (2016)
Ellouze, F., Ben Amar, N., Deratani, A.: Large ring cyclodextrins: synthesis, purification and applications. C. R. Chim. 14(10), 967–971 (2011)
Ivanov, P., Atanassov, E., Jaime, C.: Computational study on the conformations of CD38 and inclusion complexes of some lower-size large-ring cyclodextrins. J. Mol. Struct. 1056, 238–245 (2014)
Khuntawee, W., Rungrotmongkol, T., Wolschann, P., Pongsawasdi, P., Kungwan, N., Okumura, H., Hannongbua, S.: Conformation study of ε-cyclodextrin: Replica exchange molecular dynamics simulations. Carbohyd. Polym. 141, 99–105 (2016)
Crich, D.: Modern Synthetic Methods in Carbohydrate Chemistry: From Monosaccharides to Complex Glycoconjugates. Wiley, Weinheim (2013)
Alvarez-Dorta, D., León, E.I., Kennedy, A.R., Martín, A., Pérez-Martín, I., Suárez, E.: Easy access to modified cyclodextrins by an intramolecular radical approach. Angew. Chem. Int. Ed. 54(12), 3674–3678 (2015)
Cheng, G.-J., Zhang, X., Chung, L.W., Xu, L., Wu, Y.-D.: Computational organic chemistry: bridging theory and experiment in establishing the mechanisms of chemical reactions. J. Am. Chem. Soc. 137(5), 1706–1725 (2015)
Shityakov, S., Salmas, R.E., Durdagi, S., Salvador, E., Pápai, K., Yáñez-Gascón, M.J., Pérez-Sánchez, H., Puskás, I., Roewer, N., Förster, C.: Characterization, in vivo evaluation, and molecular modeling of different propofol–cyclodextrin complexes to assess their drug delivery potential at the blood–brain barrier level. J. Chem. Inf. Model. 56(10), 1914–1922 (2016)
Kirschner, K.N., Yongye, A.B., Tschampel, S.M., González-Outeiriño, J., Daniels, C.R., Foley, B.L., Woods, R.J.: GLYCAM06: a generalizable biomolecular force field. Carbohydr. J. Comput. Chem. 29(4), 622–655 (2008)
Wang, J., Wolf, R.M., Caldwell, J.W., Kollman, P.A., Case, D.A.: Development and testing of a general amber force field. J. Comput. Chem. 25(9), 1157–1174 (2004)
Devasari, N., Dora, C.P., Singh, C., Paidi, S.R., Kumar, V., Sobhia, M.E., Suresh, S.: Inclusion complex of erlotinib with sulfobutyl ether-β-cyclodextrin: preparation, characterization, in silico, in vitro and in vivo evaluation. Carbohyd. Polym. 134, 547–556 (2015)
Shityakov, S., Puskás, I., Pápai, K., Salvador, E., Roewer, N., Förster, C., Broscheit, J.-A.: Sevoflurane-sulfobutylether-β-cyclodextrin complex: preparation, characterization, cellular toxicity, molecular modeling and blood-brain barrier transport studies. Molecules 20(6), 10264–10279 (2015)
Kulkarni, A.D., Belgamwar, V.S.: Inclusion complex of chrysin with sulfobutyl ether-β-cyclodextrin (Captisol®): Preparation, characterization, molecular modelling and in vitro anticancer activity. J. Mol. Struct. 1128, 563–571 (2017)
Yildiz, Z.I., Celebioglu, A., Uyar, T.: Polymer-free electrospun nanofibers from sulfobutyl ether 7-beta-cyclodextrin (SBE 7-β-CD) inclusion complex with sulfisoxazole: fast-dissolving and enhanced water-solubility of sulfisoxazole. Int. J. Pharmaceut. 531(2), 550–558 (2017)
Altarsha, M., Ingrosso, F., Ruiz-López, M.F.: Cavity closure dynamics of peracetylated β-cyclodextrins in supercritical carbon dioxide. J. Phys. Chem. B 116(13), 3982–3990 (2012)
Bayly, C.I., Cieplak, P., Cornell, W.D., Kollman, P.A.: A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97(40), 10269–10280 (1993)
Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Merz, K.M. Jr., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., Kollman, P.A.: A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117(19), 5179–5197 (1995)
Kordopati, G.G., Tselios, T.V., Kellici, T., Merzel, F., Mavromoustakos, T., Grdadolnik, S.G., Tsivgoulis, G.M.: A novel synthetic luteinizing hormone-releasing hormone (LHRH) analogue coupled with modified β-cyclodextrin: insight into its intramolecular interactions. Biochim. Biophys. Acta 1850(1), 159–168 (2015)
Wang, R., Zhou, H., Siu, S.W., Gan, Y., Wang, Y., Ouyang, D.: Comparison of three molecular simulation approaches for cyclodextrin-ibuprofen complexation. J. Nanomater. 16(1), 267 (2015)
Sheng Cai, W., Wang, T., Zhe Liu, Y., Liu, P., Chipot, C., Guang Shao, X.: Free energy calculations for cyclodextrin inclusion complexes. Curr. Org. Chem. 15(6), 839–847 (2011)
Sangpheak, W., Khuntawee, W., Wolschann, P., Pongsawasdi, P., Rungrotmongkol, T.: Enhanced stability of a naringenin/2, 6-dimethyl β-cyclodextrin inclusion complex: molecular dynamics and free energy calculations based on MM-and QM-PBSA/GBSA. J. Mol. Graph. Model. 50, 10–15 (2014)
Rutenberg, R., Leitus, G., Fallik, E., Poverenov, E.: Discovery of a non classic host guest complexation mode in a β-cyclodextrin/propionic acid model. Chem. Commun. 52(12), 2565–2568 (2016)
Rahim, M., Madi, F., Nouar, L., Haiahem, S., Fateh, D., Khatmi, D.: β-Cyclodextrin Interaction with Edaravone: Molecular Modeling Study. Proceedings of MEST 2012: Electronic Structure Methods with Applications to Experimental Chemistry, vol. 68, pp. 269–278 (2014)
Onnainty, R.e., Schenfeld, E.M., Quevedo, M.A., Fernández, M.A., Longhi, M.R., Granero, G.E.: Characterization of the hydrochlorothiazide: β-cyclodextrin inclusion complex. Experimental and theoretical methods. J. Phys. Chem. B 117(1), 206–217 (2012)
Oda, M., Kuroda, M.: Molecular dynamics simulations of inclusion complexation of glycyrrhizic acid and cyclodextrins (1: 1) in water. J. Incl. Phenom. Macrocycl. Chem. 85(3–4), 271–279 (2016)
Nociari, M.M., Lehmann, G.L., Bay, A.E.P., Radu, R.A., Jiang, Z., Goicochea, S., Schreiner, R., Warren, J.D., Shan, J., de Beaumais, S.A.: Beta cyclodextrins bind, stabilize, and remove lipofuscin bisretinoids from retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 111(14), E1402–E1408 (2014)
Kogawa, A.C., Zoppi, A., Quevedo, M.A., Raquel, M.: Complexation between darunavir ethanolate and β-cyclodextrin experimental and theoretical studies. World J. Pharm. Pharmaceut. Sci. 298–309 (2014)
Al-Rawashdeh, N.A.F., Al-Sadeh, K.S., Al-Bitar, M.B.: Inclusion complexes of sunscreen agents with β-cyclodextrin: spectroscopic and molecular modeling studies. J. Spectrosc. 1(1), 1–11 (2013)
Mobley, D.L., Gilson, M.K.: Predicting binding free energies: frontiers and benchmarks. Ann. Rev. Biophys. 46, 531–558 (2017)
Abel, R., Wang, L., Mobley, D.L., Friesner, R.A.: A critical review of validation, blind testing, and real-world use of alchemical protein-ligand binding free energy calculations. Curr. Top. Med. Chem. 17(23), 2577–2585 (2017)
Torrie, G.M., Valleau, J.P.: Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J. Comput. Phys. 23(2), 187–199 (1977)
Wickstrom, L., He, P., Gallicchio, E., Levy, R.M.: Large scale affinity calculations of cyclodextrin host–guest complexes: understanding the role of reorganization in the molecular recognition process. J. Chem. Theory Comput. 9(7), 3136–3150 (2013)
Zhang, H., Yin, C., Yan, H., van der Spoel, D.: Evaluation of generalized born models for large scale affinity prediction of cyclodextrin host–guest complexes. J. Chem. Inform. Model. 56(10), 2080–2092 (2016)
Veselinović, A.M., Veselinović, J.B., Toropov, A.A., Toropova, A.P., Nikolić, G.M.: In silico prediction of the β-cyclodextrin complexation based on Monte Carlo method. Int. J. Pharmaceut. 495(1), 404–409 (2015)
Tan, Z., Xia, J., Zhang, B.W., Levy, R.M.: Locally weighted histogram analysis and stochastic solution for large-scale multi-state free energy estimation. J. Chem. Phys. 144(3), 034107 (2016)
Sugita, M., Hirata, F.: Predicting the binding free energy of the inclusion process of 2-hydroxypropyl-β-cyclodextrin and small molecules by means of the MM/3D-RISM method. J. Phys. Condens. Mater. 28(38), 384002 (2016)
Sancho, M.I., Andujar, S., Porasso, R.D., Enriz, R.D.: Theoretical and experimental study of inclusion complexes of β-cyclodextrins with chalcone and 2′,4′-dihydroxychalcone. J. Phys. Chem. B 120(12), 3000–3011 (2016)
Sahra, K., Dinar, K., Seridi, A., Kadri, M.: Investigation on the inclusion of diclofenac with β-cyclodextrin: a molecular modeling approach. Struct. Chem. 26(1), 61–69 (2015)
Angelova, S., Nikolova, V., Molla, N., Dudev, T.: Factors Governing the host–guest interactions between IIA/IIB group metal cations and α-cyclodextrin: a DFT/CDM study. Inorg. Chem. 56(4), 1981–1987 (2017)
Ateba, B.A., Lissouck, D., Azébazé, A., Ebelle, C.T., Nassi, A., Ngameni, E., Duportail, G., Mbazé, L., Kenfack, C.A.: Characterization of Mammea A/AA in solution and in interaction with β-cyclodextrin: UV–visible spectroscopy, cyclic voltammetry and DFT-TDDFT/MD study. J. Mol. Liq. 213, 294–303 (2016)
Henriksen, N.M., Fenley, A.T., Gilson, M.K.: Computational calorimetry: high-precision calculation of host–guest binding thermodynamics. J. Chem. Theory Comput. 11(9), 4377–4394 (2015)
Cao, R., Wu, S.: In silico properties characterization of water-soluble γ-cyclodextrin bi-capped C 60 complex: free energy and geometrical insights for stability and solubility. Carbohyd. Polym. 124, 188–195 (2015)
Tóth, G., Mohácsi, R., Rácz, Á., Rusu, A., Horváth, P., Szente, L., Béni, S., Noszál, B.: Equilibrium and structural characterization of ofloxacin–cyclodextrin complexation. J. Incl. Phenom. Macrocycl. Chem. 77(1–4), 291–300 (2013)
Shi, M., Zhang, C., Xie, Y., Xu, D.: Stereoselective inclusion mechanism of ketoprofen into β-cyclodextrin: insights from molecular dynamics simulations and free energy calculations. Theor. Chem. Acc. 133(10), 1556 (2014)
Melani, F., Pasquini, B., Caprini, C., Gotti, R., Orlandini, S., Furlanetto, S.: Combination of capillary electrophoresis, molecular modeling and NMR to study the enantioselective complexation of sulpiride with double cyclodextrin systems. J. Pharmaceut. Biomed. 114, 265–271 (2015)
Li, L., Li, X., Luo, Q., You, T.: A comprehensive study of the enantioseparation of chiral drugs by cyclodextrin using capillary electrophoresis combined with theoretical approaches. Talanta 142, 28–34 (2015)
Ghatee, M.H., Sedghamiz, T.: Chiral recognition of propranolol enantiomers by β-cyclodextrin: quantum chemical calculation and molecular dynamics simulation studies. Chem. Phys. 445, 5–13 (2014)
Amharar, Y., Grandeury, A., Sanselme, M., Petit, S., Coquerel, G.r.: A hybrid mechanism in chiral discrimination induced by crystallization of supramolecular compounds. J. Phys. Chem. B 116(20), 6027–6040 (2012)
Alvira, E.: Molecular dynamics study of the influence of solvents on the chiral discrimination of alanine enantiomers by β-cyclodextrin. Tetrahedron Asymmetr. 24(19), 1198–1206 (2013)
Alvira, E.: Influence of molecular stereochemistry on the continuum model for van der waals interaction between β-cyclodextrin and linear molecules. Curr. Phys. Chem. 3(3), 357–365 (2013)
Alvira, E.: Theoretical study of the separation of valine enantiomers by β-cyclodextrin with different solvents: a molecular mechanics and dynamics simulation. Tetrahedron Asymmetr. 26(15), 853–860 (2015)
Abou-Zeid, L.A., Hefnawy, M.: Molecular modeling study of the chiral recognition of celiprolol enantiomers by a β-cyclodextrin. Pharmaceut. Chem. J. 2(3), 16–23 (2015)
Suliman, F.O., Elbashir, A.A.: Enantiodifferentiation of chiral baclofen by β-cyclodextrin using capillary electrophoresis: a molecular modeling approach. J. Mol. Struct. 1019, 43–49 (2012)
Periasamy, R., Kothainayaki, S., Sivakumar, K.: Encapsulation of dicinnamalacetone in β-cyclodextrin: A physicochemical evaluation and molecular modeling approach on 1: 2 inclusion complex. J. Macromol. Sci. A 53(9), 546–556 (2016)
Terekhova, I., Kumeev, R., Alper, G., Chakraborty, S., Pérez-Sánchez, H., Núñez-Delicado, E.: Molecular recognition of aromatic carboxylic acids by hydroxypropyl-γ-cyclodextrin: experimental and theoretical evidence. RSC Adv. 6(55), 49567–49577 (2016)
Zhang, H., Ge, C., van der Spoel, D., Feng, W., Tan, T.: Insight into the structural deformations of beta-cyclodextrin caused by alcohol cosolvents and guest molecules. J. Phys. Chem. B 116(12), 3880–3889 (2012)
Zhang, H., Feng, W., Li, C., Lv, Y., Tan, T.: A model for the shuttle motions of puerarin and daidzin inside the cavity of β-cyclodextrin in aqueous acetic acid: insights from molecular dynamics simulations. J. Mol. Model. 18(1), 221–227 (2012)
Chandrasekaran, S., Sudha, N., Premnath, D., Enoch, I.V.: Binding of a chromen-4-one Schiff’s base with bovine serum albumin: capping with β-cyclodextrin influences the binding. J. Biomol. Struct. Dyn. 33(9), 1945–1956 (2015)
Sameena, Y., Sudha, N., Chandrasekaran, S., Enoch, I.V.: The role of encapsulation by β-cyclodextrin in the interaction of raloxifene with macromolecular targets: a study by spectroscopy and molecular modeling. J. Biol. Phys. 40(4), 347–367 (2014)
Yan, J., Wu, D., Ma, X., Wang, L., Xu, K., Li, H.: Spectral and molecular modeling studies on the influence of β-cyclodextrin and its derivatives on aripiprazole-human serum albumin binding. Carbohyd. Polym. 131, 65–74 (2015)
Tang, P., Tang, B., Wang, Q., Xu, K., Xiong, X., Li, H.: Effect of hydroxypropyl-β-cyclodextrin on the bounding of salazosulfapyridine to human serum albumin. Int. J. Biol. Macromol. 92, 105–115 (2016)
Natesan, S., Sowrirajan, C., Dhanaraj, P., Enoch, I.V.: Capping of silybin with β-cyclodextrin influences its binding with bovine serum albumin: a study by fluorescence spectroscopy and molecular modeling. Bull. Korean Chem. Soc. 35(7), 2114–2122 (2014)
Mansouri, M., Pirouzi, M., Saberi, M.R., Ghaderabad, M., Chamani, J.: Investigation on the interaction between cyclophosphamide and lysozyme in the presence of three different kind of cyclodextrins: determination of the binding mechanism by spectroscopic and molecular modeling techniques. Molecules 18(1), 789–813 (2013)
Figueiras, A., Sarraguça, J.M., Pais, A.A., Carvalho, R.A., Veiga, J.F.: The role of l-arginine in inclusion complexes of omeprazole with cyclodextrins. AAPS PharmSci Tech. 11(1), 233–240 (2010)
Sherje, A.P., Kulkarni, V., Murahari, M., Nayak, U.Y., Bhat, P., Suvarna, V., Dravyakar, B.: Inclusion complexation of etodolac with hydroxypropyl-beta-cyclodextrin and auxiliary agents: Formulation characterization and molecular modeling studies. Mol. Pharmaceut. 14(4), 1231–1242 (2017)
Sapte, S., Pore, Y.: Inclusion complexes of cefuroxime axetil with β-cyclodextrin: physicochemical characterization, molecular modeling and effect of l-arginine on complexation. J. Pharm. Anal. 6(5), 300–306 (2016)
Suvarna, V., Kajwe, A., Murahari, M., Pujar, G.V., Inturi, B.K., Sherje, A.P.: Inclusion complexes of nateglinide with HP–β–CD and l-arginine for solubility and dissolution enhancement: preparation, characterization, and molecular docking study. J. Pharm. Innov. 12(2), 168–181 (2017)
Méndez, S.G., Otero Espinar, F.J., Alvarez, A.L., Longhi, M.R., Quevedo, M.A., Zoppi, A.: Ternary complexation of benzoic acid with β-cyclodextrin and aminoacids. Experimental and theoretical studies. J. Incl. Phenom. Macrocycl. Chem. 85(1–2), 33–48 (2016). https://doi.org/10.1007/s10847-016-0603-6
Jadhav, P., Petkar, B., Pore, Y., Kulkarni, A., Burade, K.: Physicochemical and molecular modeling studies of cefixime–l-arginine–cyclodextrin ternary inclusion compounds. Carbohyd. Polym. 98(2), 1317–1325 (2013)
Barbosa, J.A.A., Zoppi, A., Quevedo, M.A., de Melo, P.N., de Medeiros, A.S.A., Streck, L., de Oliveira, A.R., Fernandes-Pedrosa, M.F., Longhi, M.R., da Silva-Júnior, A.A.: Triethanolamine stabilization of methotrexate-β-cyclodextrin interactions in ternary complexes. Int. J. Mol. Sci. 15(9), 17077–17099 (2014)
Li, L., Zhao, M., Li, W., Wang, Y., Zhang, Z., An, R., Peng, S.: Self-complexation and complexation-controlled target cancer therapy. MedChemComm 3(9), 1059–1061 (2012)
He, H., Chen, S., Zhou, J., Dou, Y., Song, L., Che, L., Zhou, X., Chen, X., Jia, Y., Zhang, J.: Cyclodextrin-derived pH-responsive nanoparticles for delivery of paclitaxel. Biomaterials 34(21), 5344–5358 (2013)
Shi, Q., Zhang, L., Liu, M., Zhang, X., Zhang, X., Xu, X., Chen, S., Li, X., Zhang, J.: Reversion of multidrug resistance by a pH-responsive cyclodextrin-derived nanomedicine in drug resistant cancer cells. Biomaterials 67, 169–182 (2015)
Xiong, Q., Zhang, M., Zhang, Z., Shen, W., Liu, L., Zhang, Q.: Anti-tumor drug delivery system based on cyclodextrin-containing pH-responsive star polymer: in vitro and in vivo evaluation. Int. J. Pharmaceut. 474(1), 232–240 (2014)
Dan, Z., Cao, H., He, X., Zeng, L., Zou, L., Shen, Q., Zhang, Z.: Biological stimuli-responsive cyclodextrin-based host–guest nanosystems for cancer therapy. Int. J. Pharmaceut. 483(1), 63–68 (2015)
Swiech, O., Mieczkowska, A., Chmurski, K., Bilewicz, R.: Intermolecular interactions between doxorubicin and β-cyclodextrin 4-methoxyphenol conjugates. J. Phys. Chem. B 116(6), 1765–1771 (2012)
Hrubý, M., Koňák, Č., Ulbrich, K.: Polymeric micellar pH-sensitive drug delivery system for doxorubicin. J. Control Release 103(1), 137–148 (2005)
Swiech, O., Majdecki, M., Debinski, A., Krzak, A., Stępkowski, T.M., Wójciuk, G., Kruszewski, M., Bilewicz, R.: Competition between self-inclusion and drug binding explains the pH dependence of the cyclodextrin drug carrier–molecular modelling and electrochemistry studies. Nanoscale 8(37), 16733–16742 (2016)
Swiech, O., Dutkiewicz, P., Wójciuk, K., Chmurski, K., Kruszewski, M., Bilewicz, R.: Cyclodextrin derivatives conjugated with aromatic moieties as pH-responsive drug carriers for anthracycline. J. Phys. Chem. B 117(43), 13444–13450 (2013)
De Sousa, F.B., Lima, A.C., Denadai, Â.M., Anconi, C.P., De Almeida, W.B., Novato, W.T., Dos Santos, H.F., Drum, C.L., Langer, R., Sinisterra, R.D.: Superstructure based on β-CD self-assembly induced by a small guest molecule. Phys. Chem. Chem. Phys. 14(6), 1934–1944 (2012)
Goh, G.B., Hulbert, B.S., Zhou, H., Brooks, C.L.: Constant pH molecular dynamics of proteins in explicit solvent with proton tautomerism. Proteins: structure, function, and bioinformatics 82(7), 1319–1331 (2014)
Swails, J.M., York, D.M., Roitberg, A.E.: Constant pH replica exchange molecular dynamics in explicit solvent using discrete protonation states: implementation, testing, and validation. J. Chem. Theory Comput. 10(3), 1341–1352 (2014)
Arthur, E.J., Brooks, C.L.: Efficient implementation of constant pH molecular dynamics on modern graphics processors. J. Comput. Chem. 37(24), 2171–2180 (2016)
Harada, A., Takashima, Y., Yamaguchi, H.: Cyclodextrin-based supramolecular polymers. Chem. Soc. Rev. 38(4), 875–882 (2009)
Zhang, H., Tan, T., Feng, W., Van Der Spoel, D.: Molecular recognition in different environments: β-cyclodextrin dimer formation in organic solvents. J. Phys. Chem. B 116(42), 12684–12693 (2012)
Zhang, H., Tan, T., Hetényi, C., Van Der Spoel, D.: Quantification of solvent contribution to the stability of noncovalent complexes. J. Chem. Theory Comput. 9(10), 4542–4551 (2013)
Zhang, H., Tan, T., Hetényi, C., Lv, Y., Van Der Spoel, D.: Cooperative binding of cyclodextrin dimers to isoflavone analogues elucidated by free energy calculations. J. Phys. Chem. C 118(13), 7163–7173 (2014)
Staelens, N., Leherte, L., Vercauteren, D.P.: Formation and structural, energetic and dynamic properties of cyclodextrin host tubes and included guest molecules. Supramol. Chem. 27(1–2), 90–109 (2015)
Raffaini, G., Ganazzoli, F.: A molecular modeling study of complex formation and self-aggregation behavior of a porphyrin-β-cyclodextrin conjugate. J. Incl. Phenom. Macrocycl Chem. 76(1–2), 213–221 (2013). https://doi.org/10.1007/s10847-012-0193-x
Liu, Y., Chipot, C., Shao, X., Cai, W.: Threading or tumbling? Insight into the self-inclusion mechanism of an altro-α-cyclodextrin derivative. J. Phys. Chem. C 118(33), 19380–19386 (2014)
Wallace, S.J., Kee, T.W., Huang, D.M.: Molecular basis of binding and stability of curcumin in diamide-linked γ-cyclodextrin dimers. J. Phys. Chem. B 117(41), 12375–12382 (2013)
Trotta, F., Zanetti, M., Cavalli, R.: Cyclodextrin-based nanosponges as drug carriers. Beilstein J. Org. Chem. 8(1), 2091–2099 (2012)
Raffaini, G., Ganazzoli, F., Mele, A., Castiglione, F.: A molecular dynamics study of cyclodextrin nanosponge models. J. Incl. Phenom. Macrocycl. Chem. 75(3–4), 263–268 (2013)
Mixcoha, E., Campos-Terán, J., Piñeiro, A.n.: Surface adsorption and bulk aggregation of cyclodextrins by computational molecular dynamics simulations as a function of temperature: α-CD vs β-CD. J. Phys. Chem. B 118(25), 6999–7011 (2014)
Takayanagi, M., Ito, S., Matsumoto, K., Nagaoka, M.: Formation of reactant complex structure for initiation reaction of lactone ring-opening polymerization by cooperation of multiple cyclodextrin. J. Phys. Chem. B 120(29), 7174–7181 (2016)
Loftsson, T., Brewster, M.E.: Pharmaceutical applications of cyclodextrins: effects on drug permeation through biological membranes. J. Pharm. Pharmacol 63(9), 1119–1135 (2011)
Loftsson, T.: Drug permeation through biomembranes: cyclodextrins and the unstirred water layer. Int. J. Pharmaceut. Sci. 67(5), 363–370 (2012)
Khuntawee, W., Wolschann, P., Rungrotmongkol, T., Wong-Ekkabut, J., Hannongbua, S.: Molecular dynamics simulations of the interaction of beta cyclodextrin with a lipid bilayer. J. Chem. Inf. Model. 55(9), 1894–1902 (2015)
Hashidzume, A., Yamaguchi, H., Harada, A.: Cyclodextrin-based molecular machines. In: Credi, A., Silvi, S., Venturi, M. (eds.) Molecular Machines and Motors, pp. 71–110. Springer, Dordrecht (2014)
Wenz, G., Han, B.-H., Müller, A.: Cyclodextrin rotaxanes and polyrotaxanes. Chem. Rev. 106(3), 782–817 (2006)
Singharoy, A., Chipot, C.: Methodology for the simulation of molecular motors at different scales. J. Phys. Chem. B 121(15), 3502–3514 (2016)
Liu, P., Chipot, C., Cai, W., Shao, X.: Unveiling the underlying mechanism for compression and decompression strokes of a molecular engine. J. Phys. Chem. C 118(23), 12562–12567 (2014)
Bruns, C.J., Stoddart, J.F.: The Nature of the Mechanical Bond: From Molecules to Machines. Wiley, Hoboken (2016)
Liu, P., Chipot, C., Shao, X., Cai, W.: Solvent-controlled shuttling in a molecular switch. J. Phys. Chem. C 116(7), 4471–4476 (2012)
Low, P.J., Marqués-González, S.: Molecular wires: an overview of the building blocks of molecular electronics. In: Kiguchi, M. (ed.) Single-Molecule Electronics, pp. 87–116. Springer, Dordrecht (2016)
Tallury, S.S., Smyth, M.B., Cakmak, E., Pasquinelli, M.A.: Molecular dynamics simulations of interactions between polyanilines in their inclusion complexes with β-cyclodextrins. J. Phys. Chem. B 116(7), 2023–2030 (2012)
Muhammad, E.F., Adnan, R., Latif, M.A.M., Rahman, M.B.A.: Theoretical investigation on insulin dimer-β-cyclodextrin interactions using docking and molecular dynamics simulation. J. Incl. Phenom. Macrocycl. Chem. 84(1–2), 1–10 (2016)
Krauland, A.H., Alonso, M.J.: Chitosan/cyclodextrin nanoparticles as macromolecular drug delivery system. Int. J. Pharmaceut. 340(1–2), 134–142 (2007)
Berhanu, W.M., Masunov, A.E.: Controlling the aggregation and rate of release in order to improve insulin formulation: Molecular dynamics study of full-length insulin amyloid oligomer models. J. Mol. Model. 18(3), 1129–1142 (2012)
Muhammad, E.F.: Docking And Molecular Dynamics Simulation Studies Of Insulin-Β-Cyclodextrin Interactions. Universiti Sains Malaysia, Gelugor (2016)
Panchal, A.: Insulin drug delivery systems: a review. Int. J. Res. Pharmaceut. Sci. 2(4), 484–492 (2016)
Gonzalez-Gaitano, G., Ramon Isasi, J., Velaz, I., Zornoza, A.: Drug carrier systems based on cyclodextrin supramolecular assemblies and polymers: present and perspectives. Curr. Pharm. Design 23(3), 411–432 (2017)
Acknowledgements
The authors gratefully acknowledge financial support from the Secretaria de Ciencia y Técnica of the Universidad Nacional de Córdoba (SECYT-UNC), the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), and the Agencia Nacional de Promoción Científica y Técnica (ANPCyT), Argentina.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Quevedo, M.A., Zoppi, A. Current trends in molecular modeling methods applied to the study of cyclodextrin complexes. J Incl Phenom Macrocycl Chem 90, 1–14 (2018). https://doi.org/10.1007/s10847-017-0763-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10847-017-0763-z