Abstract
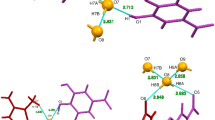
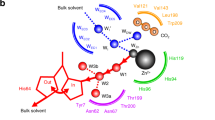
Creatininase is a key enzyme of creatinine-metabolizing pathway in mammals, and has a great potential for diagnostic application. It catalyzes the reversible conversion of creatinine to creatine. Here, we investigated its reaction mechanism with density functional theory in conjunction with the quantum cluster approach. Three reaction pathways in which several possible proton transfers assisted by either His178 or a water ligand to Zn1 (Wat2) or both were considered. DFT calculations reveal, depending on Wat2 coordination mode at Zn1, two competitive ring-opening pathways where His178 playing a central role as a proton shuttle or both His178 and Wat2 serving as a dual catalytic role as a base and an acid, respectively. Three elementary steps were proposed for the reaction: the first involves nucleophilic attack by a bridging hydroxide to the substrate and forms a gem-diolate intermediate, followed by a proton transfer from the gem-diolate to His178 (His178 protonation is a required step for efficient proton transfers). Finally, the second proton transfer from the protonated His178 or Wat2 to the amide of substrate leads to the ring opening. The first proton transfer is the rate-limiting step of the whole reaction, in consistent with previous experimental and computational studies. A detailed understanding of the reaction mechanism of the creatininase enzyme family will also be helpful for developing a biosensor for kidney function.









Similar content being viewed by others
Data Availability
All data generated or analysed during this study are included in this published article (and its supplementary information files).
References
Szulmajster J (1958) Bacterial degradation of creatinine. II. Creatinine desimidase. Biochim Biophys Acta 30(1):154
Wyss M, Kaddurah-Daouk R (2000) Creatine and creatinine metabolism. Physiol Rev 80(3):1107
Kaplan A, Szabo LL (1974) Creatinine hydrolase and creatine amidinohydrolase. II. Partial purification and properties. Mol Cell Biochem 3:17
Tsuru D, Oka I, Yoshimoto T (1976) Creatinine decomposing enzymes in Pseudomonas putida. Agric Biol Chem 40:1011
Rikitake K, Oka I, Ando M, Yoshimoto T, Tsuru D (1979) Creatinine amidohydrolase (creatininase) from Pseudomonas putida. Purification and some properties. J Biochem 86:1109
Tang TY, Wen CJ (2000) Expression of the creatininase gene from Pseudomonas putida RS65 in Escherichia coli. J Ind Microbiol 24:2
Yoshimoto T, Oka I, Tsuru D (1976) Creatine amidinohydrolase of Pseudomonas putida: Crystallization and some properties. Arch Biochem Biophys 177(2):508
Schumann J, Böhm G, Schumacher G, Rudolph R, Jaenicke R (1993) Stabilization of creatinase from Pseudomonas putida by random mutagenesis. Protein Sci 2(10):1612
Yamamoto K, Oka M (1995) Cloning of the creatinine amidohydrolase gene from Pseudomonas sp. PS-7. Biosci Biotechnol Biochem 59:1331
Beuth B, Niefind K, Schomburg D (2002) Crystallization and preliminary crystallographic analysis of creatininase from Pseudomonas putida. Acta Crystallogr D Biol Crystallogr 58(8):1356
Beuth B, Niefind K, Schomburg D (2003) Crystal structure of creatininase from Pseudomonas putida: a novel fold and a case of convergent evolution. J Mol Biol 332(1):287
Ito K, Kanada N, Inoue T, Furukawa K, Yamashita K, Tanaka N, Nakamura KT, Nishiya Y, Sogabe A, Yoshimoto T (2002) Preliminary crystallographic studies of the creatinine amidohydrolase from Pseudomonas putida. Acta Crystallogr Sect D 58:2180
Yamashita K, Nakajima Y, Matsushita H, Nishiya Y, Yamazawa R, Wu YF, Matsubara F, Oyama H, Ito K, Yoshimoto T (2010) Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase. J Mol Biol 396(4):1081
Yoshimoto T, Tanaka N, Kanada N, Inoue T, Nakajima Y, Haratake M, Nakamura KT, Xu Y, Ito K (2004) Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism. J Mol Biol 337(2):399
Jitonnom J, Mujika JI, van der Kamp MW, Mulholland AJ (2017) Quantum mechanics/molecular mechanics simulations identify the ring-opening mechanism of creatininase. Biochemistry 56(48):6377
Seibert CM, Raushel FM (2005) Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44(17):6383
Liao R-Z, Yu J-G, Frank M, Raushel, Himo F (2008) Theoretical investigation of the reaction mechanism of the dinuclear zinc enzyme dihydroorotase. Chem Eur J 14:4287
Liao R-Z, Himo F, Yu J-G, Liu R-Z (2010) Dipeptide hydrolysis by the dinuclear zinc enzyme human renal dipeptidase: Mechanistic insights from DFT calculations. J Inorg Biochem 104(1):37
Liao R-Z, Yu J-G, Himo F (2009) Reaction Mechanism of the Dinuclear Zinc Enzyme N-Acyl-l-homoserine Lactone Hydrolase: A Quantum Chemical Study. Inorg Chem 48(4):1442
Krivitskaya AV, Khrenova MG, Nemukhin AV (2021) Two Sides of Quantum-Based Modeling of Enzyme-Catalyzed Reactions: Mechanistic and Electronic Structure Aspects of the Hydrolysis by Glutamate Carboxypeptidase. Molecules 26(20):6280
Reidl CT, Mascarenhas R, Mohammad TSH, Lutz MR Jr, Thomas PW, Fast W, Liu D, Becker DP (2021) Cyclobutanone Inhibitor of Cobalt-Functionalized Metallo-γ-Lactonase AiiA with Cyclobutanone Ring Opening in the Active Site. ACS omega 6(21):13567
Liao R-Z, Yu J-G, Himo F (2010) Reaction Mechanism of the Trinuclear Zinc Enzyme Phospholipase C: A Density Functional Theory Study. J Phys Chem B 114(7):2533
Zhu X, Barman A, Ozbil M, Zhang T, Li S, Prabhakar R (2012) Mechanism of peptide hydrolysis by co-catalytic metal centers containing leucine aminopeptidase enzyme: a DFT approach. J Biol Inorg Chem 17(2):209
Manta B, Raushel FM, Himo F (2014) Reaction mechanism of zinc-dependent cytosine deaminase from Escherichia coli: a quantum-chemical study. J Phys Chem B 118(21):5644
Jitonnom J, Sattayanon C, Kungwan N, Hannongbua S (2015) A DFT study of the unusual substrate-assisted mechanism of Serratia marcescens chitinase B reveals the role of solvent and mutational effect on catalysis. J Mol Graph Model 56:53
Jitonnom J, Hannongbua S (2018) Theoretical study of the arabinan hydrolysis by an inverting GH43 arabinanase. Mol Simul 44(8):631
Fernandes HS, Teixeira CSS, Sousa SF, Cerqueira NMFSA (2019) Formation of unstable and very reactive chemical species catalyzed by metalloenzymes: a mechanistic overview.Molecules24(13)
Jitonnom J, Lee VS, Nimmanpipug P, Rowlands HA, Mulholland AJ (2011) Quantum mechanics/molecular mechanics modeling of substrate-assisted catalysis in family 18 chitinases: conformational changes and the role of Asp142 in catalysis in ChiB. Biochemistry 50(21):4697
Himo F (2017) Recent Trends in Quantum Chemical Modeling of Enzymatic Reactions. J Am Chem Soc 139(20):6780
Siegbahn PEM, Himo F (2011) The quantum chemical cluster approach for modeling enzyme reactions. Wiley Interdisciplinary Reviews: Computational Molecular Science 1(3):323
Siegbahn PE, Himo F (2009) Recent developments of the quantum chemical cluster approach for modeling enzyme reactions. J Biol Inorg Chem 14(5):643
Ahmadi S, Barrios Herrera L, Chehelamirani M, Hostaš J, Jalife S, Salahub DR (2018) Multiscale modeling of enzymes: QM-cluster, QM/MM, and QM/MM/MD: A tutorial review. Int J Quantum Chem 118(9):e25558
Mulliken RS (1955) Electronic population analysis on LCAO–MO molecular wave functions. I J Chem Phys 23(10):1833
Cossi M, Rega N, Scalmani G, Barone V (2003) Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J Comput Chem 24(6):669
Grimme S, Antony J, Ehrlich S, Krieg H (2010) A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys 132(15):154104
Grimme S, Ehrlich S, Goerigk L (2011) Effect of the damping function in dispersion corrected density functional theory. J Comput Chem 32(7):1456
Brás NF, Fernandes PA, Ramos MJ (2014) QM/MM study and MD simulations on the hypertension regulator angiotensin-converting enzyme. ACS Catal 4(8):2587
Lonsdale R, Harvey JN, Mulholland AJ (2010) Inclusion of dispersion effects significantly improves accuracy of calculated reaction barriers for cytochrome P450 catalyzed reactions. J Phys Chem Lett 1(21):3232
Lonsdale R, Harvey JN, Mulholland AJ (2012) Effects of dispersion in density functional based quantum mechanical/molecular mechanical calculations on cytochrome P450 catalyzed reactions. J Chem Theory Comput 8(11):4637
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, Caricato M, Marenich AV, Bloino J, Janesko BG, Gomperts R, Mennucci B, Hratchian HP, Ortiz JV, Izmaylov AF, Sonnenberg JL, Williams, Ding F, Lipparini F, Egidi F, Goings J, Peng B, Petrone A, Henderson T, Ranasinghe D, Zakrzewski VG, Gao J, Rega N, Zheng G, Liang W, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Throssell K, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark MJ, Heyd JJ, Brothers EN, Kudin KN, Staroverov VN, Keith TA, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Millam JM, Klene M, Adamo C, Cammi R, Ochterski JW, Martin RL, Morokuma K, Farkas O, Foresman JB, Fox DJ (2009) Gaussian 09 Rev. A.02. Wallingford, CT;
Kim H, Lipscomb WN (1993) Differentiation and identification of the two catalytic metal binding sites in bovine lens leucine aminopeptidase by x-ray crystallography. Proc Natl Acad Sci USA 90(11):5006
Rowlett RS, Tu C, McKay MM, Preiss JR, Loomis RJ, Hicks KA, Marchione RJ, Strong JA, Donovan GS Jr, Chamberlin JE (2002) Kinetic characterization of wild-type and proton transfer-impaired variants of beta-carbonic anhydrase from Arabidopsis thaliana. Arch Biochem Biophys 404(2):197
Acknowledgements
Financial supports by the Thailand Research Fund and the University of Phayao (Grants RSA6280104 and FF64-RIB002) are gratefully acknowledged. The School of Science, University of Phayao (Grant PBTSC65006) is also acknowledged. The authors acknowledge National e-Science Infrastructure Consortium for providing computing resources that have partly contributed to the research results reported within this paper (www.e-science.in.th).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Meelua, W., Oláh, J. & Jitonnom, J. Role of water coordination at zinc binding site and its catalytic pathway of dizinc creatininase: insights from quantum cluster approach. J Comput Aided Mol Des 36, 279–289 (2022). https://doi.org/10.1007/s10822-022-00451-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-022-00451-8