Abstract
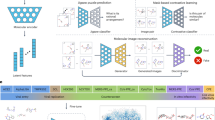
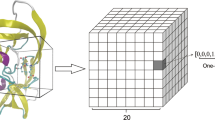
Drug Design Data Resource (D3R) Grand Challenge 4 (GC4) offered a unique opportunity for designing and testing novel methodology for accurate docking and affinity prediction of ligands in an open and blinded manner. We participated in the beta-secretase 1 (BACE) Subchallenge which is comprised of cross-docking and redocking of 20 macrocyclic ligands to BACE and predicting binding affinity for 154 macrocyclic ligands. For this challenge, we developed machine learning models trained specifically on BACE. We developed a deep neural network (DNN) model that used a combination of both structure and ligand-based features that outperformed simpler machine learning models. According to the results released by D3R, we achieved a Spearman's rank correlation coefficient of 0.43(7) for predicting the affinity of 154 ligands. We describe the formulation of our machine learning strategy in detail. We compared the performance of DNN with linear regression, random forest, and support vector machines using ligand-based, structure-based, and combining both ligand and structure-based features. We compared different structures for our DNN and found that performance was highly dependent on fine optimization of the L2 regularization hyperparameter, alpha. We also developed a novel metric of ligand three-dimensional similarity inspired by crystallographic difference density maps to match ligands without crystal structures to similar ligands with known crystal structures. This report demonstrates that detailed parameterization, careful data training and implementation, and extensive feature analysis are necessary to obtain strong performance with more complex machine learning methods. Post hoc analysis shows that scoring functions based only on ligand features are competitive with those also using structural features. Our DNN approach tied for fifth in predicting BACE-ligand binding affinities.









Similar content being viewed by others
References
Gathiaka S, Liu S, Chiu M et al (2016) D3R grand challenge 2015: evaluation of protein-ligand pose and affinity predictions. J Comput Aided Mol Des 30:651–668. https://doi.org/10.1007/s10822-016-9946-8
Gaieb Z, Liu S, Gathiaka S et al (2018) D3R Grand Challenge 2: blind prediction of protein–ligand poses, affinity rankings, and relative binding free energies. J Comput Aided Mol Des 32:1–20. https://doi.org/10.1007/s10822-017-0088-4
Gaieb Z, Parks CD, Chiu M et al (2019) D3R Grand Challenge 3: blind prediction of protein–ligand poses and affinity rankings. J Comput Aided Mol Des 33:1–18. https://doi.org/10.1007/s10822-018-0180-4
Bajorath J (2015) Computer-aided drug discovery. F Res 4:630. https://doi.org/10.12688/f1000research.6653.1
Ferreira LG, dos Santos RN, Oliva G, Andricopulo AD (2015) Molecular docking and structure-based drug design strategies. Molecules 20:13384–13421. https://doi.org/10.3390/molecules200713384
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Ravindranath PA, Forli S, Goodsell DS et al (2015) AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLoS Comput Biol 11:1–28. https://doi.org/10.1371/journal.pcbi.1004586
Trott O, Olson AJ (2009) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Friesner RA, Banks JL, Murphy RB et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749. https://doi.org/10.1021/jm0306430
Taylor R, Cole J, Cosgrove D et al (2012) Development and validation of an improved algorithm for overlaying flexible molecules. J Comput Aided Mol Des 26:451–472. https://doi.org/10.1007/s10822-012-9573-y
Wang R, Lai L, Wang S (2002) Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des 16:11–26
Khamis MA, Khamis MAM (2015) Machine learning in computational docking. Artif Intell Med 63:135–152
Lima AN, Philot EA, Trossini GHG et al (2016) Use of machine learning approaches for novel drug discovery. Expert Opin Drug Discov 11:225–239. https://doi.org/10.1517/17460441.2016.1146250
Sanchez-Lengeling B, Aspuru-Guzik A (2018) Inverse molecular design using machine learning: generative models for matter engineering. Science 361:360–365. https://doi.org/10.1126/science.aat2663
Abadi M, Agarwal A, Barham P et al (2016) TensorFlow: large-scale machine learning on heterogeneous distributed systems. ArXiv160304467 Cs
Pedregosa F, Varoquaux G, Gramfort A et al (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830
Berman HM, Westbrook J, Feng Z et al (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Liu Z, Su M, Han L et al (2017) Forging the basis for developing protein–ligand interaction scoring functions. Acc Chem Res 50:302–309. https://doi.org/10.1021/acs.accounts.6b00491
Ballester PJ, Mitchell JBO (2010) A machine learning approach to predicting protein–ligand binding affinity with applications to molecular docking. Bioinform Oxf Engl 26:1169–1175. https://doi.org/10.1093/bioinformatics/btq112
Jiménez J, Škalič M, Martínez-Rosell G, De Fabritiis G (2018) KDEEP: protein–ligand absolute binding affinity prediction via 3D-convolutional neural networks. J Chem Inf Model 58:287–296. https://doi.org/10.1021/acs.jcim.7b00650
Quiroga R, Villarreal MA (2016) Vinardo: a scoring function based on Autodock Vina improves scoring, docking, and virtual screening. PLoS ONE 11:1–18. https://doi.org/10.1371/journal.pone.0155183
Koes DR, Baumgartner MP, Camacho CJ (2013) Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J Chem Inf Model 53:1893–1904. https://doi.org/10.1021/ci300604z
Li H, Leung K-S, Wong M-H, Ballester PJ (2015) Improving AutoDock Vina using random forest: the growing accuracy of binding affinity prediction by the effective exploitation of larger data sets. Mol Inform 34:115–126. https://doi.org/10.1002/minf.201400132
Ashtawy HM, Mahapatra NR (2012) A comparative assessment of ranking accuracies of conventional and machine-learning-based scoring functions for protein–ligand binding affinity prediction. IEEE/ACM Trans Comput Biol Bioinform 9:1301–1313. https://doi.org/10.1109/TCBB.2012.36
Cang Z, Mu L, Wei G-W (2018) Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLOS Comput Biol 14:e1005929. https://doi.org/10.1371/journal.pcbi.1005929
Durrant JD, McCammon JA (2011) NNScore 2.0: a neural-network receptor-ligand scoring function. J Chem Inf Model 51:2897–2903. https://doi.org/10.1021/ci2003889
Smith JS, Isayev O, Roitberg AE (2017) ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem Sci 8:3192–3203. https://doi.org/10.1039/C6SC05720A
Sander T, Freyss J, von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55:460–473. https://doi.org/10.1021/ci500588j
O’Boyle NM, Banck M, James CA et al (2011) Open Babel: an open chemical toolbox. J Cheminformatics 3:33. https://doi.org/10.1186/1758-2946-3-33
Alvarez S (2013) A cartography of the van der Waals territories. Dalton Trans 42:8617–8636. https://doi.org/10.1039/C3DT50599E
Schrödinger, LLC PYMOL, The PyMOL Molecular Graphics System, Version 2.0
Vassar R, Bennett BD, Babu-Khan S et al (1999) Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286:735–741
Acknowledgements
This work was funded by NSF CAREER MCB 1833181.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, B., Ng, HL. Deep neural network affinity model for BACE inhibitors in D3R Grand Challenge 4. J Comput Aided Mol Des 34, 201–217 (2020). https://doi.org/10.1007/s10822-019-00275-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00275-z