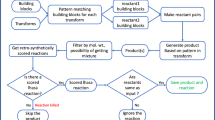
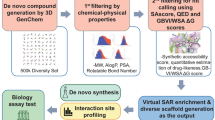
Abstract
Generative topographic mapping was used to investigate the possibility to diversify the in-house compounds collection of Boehringer Ingelheim (BI). For this purpose, a 2D map covering the relevant chemical space was trained, and the BI compound library was compared to the Aldrich-Market Select (AMS) database of more than 8M purchasable compounds. In order to discover new (sub)structures, the “AutoZoom” tool was developed and applied in order to analyze chemotypes of molecules residing in heavily populated zones of a map and to extract the corresponding maximum common substructures. A set of 401K new structures from the AMS database was retrieved and checked for drug-likeness and biological activity.








Similar content being viewed by others
Abbreviations
- GTM:
-
Generative topographic mapping
- FS:
-
Frame set
- LLh:
-
Likelihood
- AD:
-
Applicability domain
- RBF:
-
Radial basis function
- DB:
-
Database
- AMS:
-
Aldrich Market Select
- BI:
-
Boehringer Ingelheim
- MCS:
-
Maximum common substructure
References
Aladinskiy V, Sanchez-Lengeling B, Aspuru-Guzik A et al (2018) Reinforced adversarial neural computer for de novo molecular design. J Chem Inf Model 58:1194–1204. https://doi.org/10.1021/acs.jcim.7b00690
Kang S, Cho K (2019) Conditional molecular design with deep generative models. J Chem Inf Model 59:43–52. https://doi.org/10.1021/acs.jcim.8b00263
Schneider P, Schneider G (2016) De novo design at the edge of chaos: miniperspective. J Med Chem 59:4077–4086
Sattarov B, Baskin II, Horvath D et al (2019) De novo molecular design by combining deep autoencoder recurrent neural networks with generative topographic mapping. J Chem Inf Model 59:1182–1196. https://doi.org/10.1021/acs.jcim.8b00751
Ruddigkeit L, Van Deursen R, Blum LC, Reymond JL (2012) Enumeration of 166 billion organic small molecules in the chemical universe database GDB-17. J Chem Inf Model 52:2864–2875. https://doi.org/10.1021/ci300415d
Chang J-W, Jin D-S (2003) A new cell-based clustering method for large, high-dimensional data in data mining applications. In: Proceedings of the 2002 ACM symposium on Applied computing. ACM, p 503
Medina-Franco JL, Maggiora GM, Giulianotti MA et al (2007) A similarity-based data-fusion approach to the visual characterization and comparison of compound databases. Chem Biol Drug Des 70:393–412. https://doi.org/10.1111/j.1747-0285.2007.00579.x
Akella LB, DeCaprio D (2010) Cheminformatics approaches to analyze diversity in compound screening libraries. Curr Opin Chem Biol 14:325–330
Bernard P, Golbraikh A, Kireev D et al (1998) Comparison of chemical databases: analysis of molecular diversity with self organising maps (SOM). Analusis 26:333–341. https://doi.org/10.1051/analusis:1998182
Kireeva N, Baskin II, Gaspar HA et al (2012) Generative topographic mapping (GTM): universal tool for data visualization, structure-activity modeling and dataset comparison. Mol Inform 31:301–312. https://doi.org/10.1002/minf.201100163
Gaspar HA, Baskin II, Marcou G et al (2015) GTM-based QSAR models and their applicability domains. Mol Inform 34:348–356. https://doi.org/10.1002/minf.201400153
Lin A, Horvath D, Afonina V et al (2018) Mapping of the available chemical space versus the chemical universe of lead-like compounds. ChemMedChem 13:540–554. https://doi.org/10.1002/cmdc.201700561
Tino P, Nabney I (2002) Hierarchical GTM: constructing localized nonlinear projection manifolds in a principled way. IEEE Trans Pattern Anal Mach Intell 24:639–656. https://doi.org/10.1109/34.1000238
Lin A, Horvath D, Marcou G et al (2019) Multi-task generative topographic mapping in virtual screening. J Comput Aided Mol Des 33:331–343. https://doi.org/10.1007/s10822-019-00188-x
Casciuc I, Zabolotna Y, Horvath D et al (2019) Virtual screening with generative topographic maps: how many maps are required? J Chem Inf Model 59:564–572. https://doi.org/10.1021/acs.jcim.8b00650
ChemAxon Standardizer. https://docs.chemaxon.com/display/docs/Standardizer. Accessed 1 Feb 2019
ChemAxon JChem. https://chemaxon.com/products/jchem-engines. Accessed 1 Feb 2019
Bishop CM, Svensén M, Williams CKI (1998) GTM: the generative topographic mapping. Neural Comput 10:215–234. https://doi.org/10.1162/089976698300017953
Sidorov P, Viira B, Davioud-Charvet E et al (2017) QSAR modeling and chemical space analysis of antimalarial compounds. J Comput Aided Mol Des 31:441–451. https://doi.org/10.1007/s10822-017-0019-4
Monev V (2004) Introduction to similarity searching in chemistry *. Match-Commun Math Comput Chem 51:7–38
(2019) RDKit: Open-source cheminformatics. http://www.rdkit.org. Accessed 1 Feb 2019
Rogers D, Hahn M (2010) Extended-connectivity fingerprints. J Chem Inf Model 50:742–754. https://doi.org/10.1021/ci100050t
Gaspar HA, Baskin II, Marcou G et al (2015) Chemical data visualization and analysis with incremental generative topographic mapping: big data challenge. J Chem Inf Model 55:84–94. https://doi.org/10.1021/ci500575y
Sidorov P, Gaspar H, Marcou G et al (2015) Mappability of drug-like space: towards a polypharmacologically competent map of drug-relevant compounds. J Comput Aided Mol Des 29:1087–1108. https://doi.org/10.1007/s10822-015-9882-z
Volochnyuk DM, Ryabukhin SV, Moroz YS et al (2019) Evolution of commercially available compounds for HTS. Drug Discov Today 24:390–402. https://doi.org/10.1016/j.drudis.2018.10.016
Dauber-Osguthorpe P, Roberts VA, Osguthorpe DJ et al (1988) Structure and energetics of ligand binding to proteins: escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins Struct Funct Bioinform 4:31–47. https://doi.org/10.1002/prot.340040106
Ruggiu F, Marcou G, Varnek A, Horvath D (2010) ISIDA property-labelled fragment descriptors. Mol Inform 29:855–868. https://doi.org/10.1002/minf.201000099
Marcou G, Solov’ev VP, Horvath D, Varnek A (2017) ISIDA fragmentor—user manual
Horvath D, Brown J, Marcou G, Varnek A (2014) An evolutionary optimizer of libsvm models. Challenges 5:450–472
Klimenko K, Marcou G, Horvath D, Varnek A (2016) Chemical space mapping and structure-activity analysis of the ChEMBL antiviral compound set. J Chem Inf Model 56:1438–1454. https://doi.org/10.1021/acs.jcim.6b00192
Hariharan R, Janakiraman A, Nilakantan R et al (2011) MultiMCS: a fast algorithm for the maximum common substructure problem on multiple molecules. J Chem Inf Model 51:788–806. https://doi.org/10.1021/ci100297y
Oliphant TE (2006) A guide to NumPy. Tregol Publishing, USA
Oliphant TE (2007) Python for scientific computing. Comput Sci Eng 9:10–20. https://doi.org/10.1109/MCSE.2007.58
Inc. PT (2015) Collaborative data science. In: Plotly Technol. Inc. https://plot.ly. Accessed 1 Feb 2019
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2012) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 64:4–17. https://doi.org/10.1016/j.addr.2012.09.019
Brenk R, Schipani A, James D et al (2008) Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem Chem Enabling Drug Discov 3:435–444
Baell JB, Holloway GA (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53:2719–2740
Doveston RG, Tosatti P, Dow M et al (2015) A unified lead-oriented synthesis of over fifty molecular scaffolds. Org Biomol Chem 13:859–865
Jadhav A, Ferreira RS, Klumpp C et al (2009) Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J Med Chem 53:37–51
Gaulton A, Hersey A, Nowotka ML et al (2017) The ChEMBL database in 2017. Nucleic Acids Res 45:D945–D954. https://doi.org/10.1093/nar/gkw1074
Acknowledgements
The authors thank Boehringer Ingelheim Pharma GmbH & Co KG for the provided data.
Funding
The project leading to this article has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Grant Agreement No. 676434, “Big Data in Chemistry” (“BIGCHEM”, http://bigchem.eu).
Author information
Authors and Affiliations
Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lin, A., Beck, B., Horvath, D. et al. Diversifying chemical libraries with generative topographic mapping. J Comput Aided Mol Des 34, 805–815 (2020). https://doi.org/10.1007/s10822-019-00215-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00215-x