Abstract
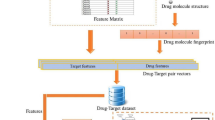
Feature selection is commonly used as a preprocessing step to machine learning for improving learning performance, lowering computational complexity and facilitating model interpretation. This paper proposes the application of boosting feature selection to improve the classification performance of standard feature selection algorithms evaluated for the prediction of P-gp inhibitors and substrates. Two well-known classification algorithms, decision trees and support vector machines, were used to classify the chemical compounds. The experimental results showed better performance for boosting feature selection with respect to the standard feature selection algorithms while maintaining the capability for feature reduction.







Similar content being viewed by others
References
Sharom F (1997) The P-glycoprotein efflux pump: how does it transport drugs? J Membr Biol 160(3):161–175
Kim RB, Fromm MF, Wandel C, Leake B, Wood AJ, Roden DM, Wilkinson GR (1998) The drug transporter P-glycoprotein limits oral absorption and brain entry of HIV-1 protease inhibitors. J Clin Invest 101(2):289–294
Fromm M (2000) P-glycoprotein: a defense mechanism limiting oral bioavailability and CNS accumulation of drugs. Int J Clin Pharmacol Ther 38(2):69–74
Marzolini C, Paus E, Buclin T, Kim RB (2004) Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance. Clin Pharmacol Ther 75(1):13–33
Szakács G, Chen GK, Gottesman MM (2004) The molecular mysteries underlying P-glycoprotein-mediated multidrug resistance. Cancer Biol Ther 3(4):382–384
Kartner N, Ling V (1983) Cell surface P-glycoprotein associated with multidrug resistance in mammalian cell lines. Cell surface P-glycoprotein associated with multidrug resistance in mammalian cell lines 221(4617):1285–1288
Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM (2006) Targeting multidrug resistance in cancer. Nat Rev Drug Discov 5(3):219
Chen L, Li Y, Zhao Q, Peng H, Hou T (2011) ADME evaluation in drug discovery. 10. Predictions of P-glycoprotein inhibitors using recursive partitioning and naive bayesian classification techniques. Mol Pharm 8(3):889–900. https://doi.org/10.1021/mp100465q
Xue Y, Yap CW, Sun LZ, Cao ZW, Wang JF, Chen YZ (2004) Prediction of P-glycoprotein substrates by a support vector machine approach. J Chem Inf Comput Sci 44(4):1497–1505. https://doi.org/10.1021/ci049971e
Huang J, Ma G, Muhammad I, Cheng Y (2007) Identifying P-glycoprotein substrates using a support vector machine optimized by a particle swarm. J Chem Inf Model 47(4):1638–1647. https://doi.org/10.1021/ci700083n
Cerqueira Lima P, Golbraikh A, Oloff S, Xiao Y, Tropsha A (2006) Combinatorial QSAR modeling of P-glycoprotein substrates. J Chem Inf Model 46(3):1245–1254
Wang Z, Chen Y, Liang H, Bender A, Glen RC, Yan A (2011) P-glycoprotein substrate models using support vector machines based on a comprehensive data set. J Chem Inf Model 51(6):1447–1456. https://doi.org/10.1021/ci2001583
Cianchetta G, Singleton RW, Zhang M, Wildgoose M, Giesing D, Fravolini A, Cruciani G, Vaz RJ (2005) A pharmacophore hypothesis for P-glycoprotein substrate recognition using GRIND-based 3D-QSAR. J Med Chem 48(8):2927–2935
Crivori P, Reinach B, Pezzetta D, Poggesi I (2006) Computational models for identifying potential P-glycoprotein substrates and inhibitors. Computational models for identifying potential P-glycoprotein substrates and inhibitors 3(1):33–44. https://doi.org/10.1021/mp050071a
Gombar VK, Polli JW, Humphreys JE, Wring SA, Serabjit-Singh CS (2004) Predicting P-glycoprotein substrates by a quantitative structure–activity relationship model. J Pharm Sci 93(4):957–968
Broccatelli F, Carosati E, Neri A, Frosini M, Goracci L, Oprea TI, Cruciani G (2011) A novel approach for predicting P-glycoprotein (ABCB1) inhibition using molecular interaction fields. J Med Chem 54(6):1740–1751
Poongavanam V, Haider N, Ecker GF (2012) Fingerprint-based in silico models for the prediction of P-glycoprotein substrates and inhibitors. Bioorg Med Chem 20(18):5388–5395
Veltri D, Kamath U, Shehu A (2014) A novel method to improve recognition of antimicrobial peptides through distal sequence-based features. In: Bioinformatics and biomedicine (BIBM), 2014 IEEE international conference on IEEE, Belfast, pp 371–378
Pérez-Rodríguez J, de Haro-García A, del Castillo JAR, García-Pedrajas N (2018) A general framework for boosting feature subset selection algorithms. Inf Fusion 44:147–175
RDKit: Open-Source Cheminformatics Software (Release 2017.03.1, 2017). http://www.rdkit.org/. Accessed 2 Sept 2017
Daylight Chemical Information Systems, Inc. http://www.daylight.com/. Accessed 2 Jan 2018
G L Fingerprints in the RDKit. http://www.rdkit.org/UGM/2012/Landrum_RDKit_UGM.Fingerprints.Final.pptx.pdf. Accessed 2 Jan 2018
Liu X, Wang H (2005) A discretization algorithm based on a heterogeneity criterion. IEEE Trans Knowl Data Eng 17(9):1166–1173. https://doi.org/10.1109/TKDE.2005.135
Goodarzi M, Dejaegher B, Heyden YV (2012) Feature selection methods in QSAR studies. J AOAC Int 95(3):636–651
Guan S-U, Liu J, Qi Y (2004) An incremental approach to contribution-based feature selection. Int J Intell Syst 13(1):15. https://doi.org/10.1515/JISYS.2004.13.1.15
Sivagaminathan RK, Ramakrishnan S (2007) A hybrid approach for feature subset selection using neural networks and ant colony optimization. Expert Syst Appl 33(1):49–60. https://doi.org/10.1016/j.eswa.2006.04.010
Newby D, Freitas AA, Ghafourian T (2013) Pre-processing feature selection for improved C&RT models for oral absorption. J Chem Inf Model 53(10):2730–2742. https://doi.org/10.1021/ci400378j
Peralta B, Soto A (2014) Embedded local feature selection within mixture of experts. Inf Sci 269:176–187
Weston J, Pérez-Cruz F, Bousquet O, Chapelle O, Elisseeff A, Schölkopf B (2003) Feature selection and transduction for prediction of molecular bioactivity for drug design. Bioinformatics 19(6):764–771. https://doi.org/10.1093/bioinformatics/btg054
Liu Y (2004) A comparative study on feature selection methods for drug discovery. J Chem Inf Comput Sci 44(5):1823–1828. https://doi.org/10.1021/ci049875d
Demel MA, Janecek AGK, Gansterer WN, Ecker GF (2009) Comparison of contemporary feature selection algorithms: application to the classification of ABC-transporter substrates. QSAR Comb Sci 28(10):1087–1091. https://doi.org/10.1002/qsar.200860191
Wassermann AM, Nisius B, Vogt M, Bajorath J (2010) Identification of descriptors capturing compound class-specific features by mutual information analysis. J Chem Inf Model 50(11):1935–1940. https://doi.org/10.1021/ci100319n
Godden JW, Bajorath J (2003) An information-theoretic approach to descriptor selection for database profiling and QSAR modeling. QSAR Comb Sci 22(5):487–497. https://doi.org/10.1002/qsar.200310001 doi
Whitley DC, Ford MG, Livingstone DJ (2000) Unsupervised forward selection: a method for eliminating redundant variables. J Chem Inf Comput Sci 40(5):1160–1168. https://doi.org/10.1021/ci000384c
Salt DW, Maccari L, Botta M, Ford MG (2004) Variable selection and specification of robust QSAR models from multicollinear data: arylpiperazinyl derivatives with affinity and selectivity for α2-adrenoceptors. J Comput Aided Mol Des 18(7):495–509. https://doi.org/10.1007/s10822-004-5203-7
Roy K, Popelier PLA (2008) Exploring predictive QSAR models for hepatocyte toxicity of phenols using QTMS descriptors. Bioorg Med Chem 18(8):2604–2609. https://doi.org/10.1016/j.bmcl.2008.03.035
Roy K, Ghosh G (2005) QSTR with extended topochemical atom indices. Part 5: Modeling of the acute toxicity of phenylsulfonyl carboxylates to Vibrio fischeri using genetic function approximation. Bioorg Med Chem 13(4):1185–1194. https://doi.org/10.1016/j.bmc.2004.11.014
Bhattacharya P, Roy K (2005) QSAR of adenosine A3 receptor antagonist 1,2,4-triazolo[4,3-a]quinoxalin-1-one derivatives using chemometric tools. Bioorg Med Chem 15(16):3737–3743. https://doi.org/10.1016/j.bmcl.2005.05.051
Yu L, Liu H (2004) Efficient feature selection via analysis of relevance and redundancy. J Mach Learn Res 5(Oct):1205–1224
Mitra P, Murthy C, Pal SK (2002) Unsupervised feature selection using feature similarity. IEEE Trans Pattern Anal Mach 24(3):301–312
Mahobia NK, Patel RD, Sheikh NW, Singh SK, Mishra A, Dhardubey R (2010) Validation method used in quantitative structure activity relationship. Der Pharma Chem 2(5):260–271
Golbraikh A, Tropsha A (2002) Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. J Comput Aided Mol Des 16(5):357–369. https://doi.org/10.1023/a:1020869118689
Schrevens E, Cornell J (1993) Design and analysis of mixture systems: applications in hydroponic, plant nutrition research. In: Optimization of plant nutrition. Springer, Berlin, pp 179–186
Snee RD (1977) Validation of regression models: methods and examples. Technometrics 19(4):415–428
Daszykowski M, Walczak B, Massart DL (2002) Representative subset selection. Anal Chim Acta 468(1):91–103. https://doi.org/10.1016/S0003-2670(02)00651-7
Bowden GJ, Maier HR, Dandy GC (2002) Optimal division of data for neural network models in water resources applications. Water Resour Res 38(2):2-1
May RJ, Maier HR, Dandy GC (2010) Data splitting for artificial neural networks using SOM-based stratified sampling. Neural Netw 23(2):283–294
Olsson IM, Gottfries J, Wold S (2004) Controlling coverage of D-optimal onion designs and selections. J Chemometr 18(12):548–557
Eriksson L, Arnhold T, Beck B, Fox T, Johansson E, Kriegl JM (2004) Onion design and its application to a pharmaceutical QSAR problem. J Chemometr 18(3–4):188–202. https://doi.org/10.1002/cem.854
Puzyn T, Mostrag-Szlichtyng A, Gajewicz A, Skrzyński M, Worth AP (2011) Investigating the influence of data splitting on the predictive ability of QSAR/QSPR models. Struct Chem 22(4):795–804
Gütlein M, Helma C, Karwath A, Kramer S (2013) A large-scale empirical evaluation of cross-validation and external test set validation in (Q) SAR. Mol Inf 32(5–6):516–528
Varmuza K, Filzmoser P, Hilchenbach M, Krüger H, Silén J (2014) KNN classification—evaluated by repeated double cross validation: recognition of minerals relevant for comet dust. Chemometr Intell Lab Syst 138:64–71. https://doi.org/10.1016/j.chemolab.2014.07.011
Ishibuchi H, Nojima Y (2013) Repeated double cross-validation for choosing a single solution in evolutionary multi-objective fuzzy classifier design. Knowl Based Syst 54:22–31
Filzmoser P, Liebmann B, Varmuza K (2009) Repeated double cross validation. J Chemometr 23(4):160–171
Luque Ruiz I, Gómez Nieto M (2018) A new data representation based on relative measurements and fingerprint patterns for the development of QSAR regression models. Chemometr Intell Lab Syst 176:53–65. https://doi.org/10.1016/j.chemolab.2018.03.007
Witten IH, Frank E, Hall MA, Pal CJ (2016) Data mining: practical machine learning tools and techniques. Morgan Kaufmann, Burlington
Bradley AP (1997) The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit 30(7):1145–1159
Cerruela García G, García-Pedrajas N, Luque Ruiz I, Gómez-Nieto M (2018) Molecular activity prediction by means of supervised subspace projection based ensembles of classifiers. SAR QSAR Environ Res 29(3):187–212. https://doi.org/10.1080/1062936X.2017.1423376
Freund Y, Schapire RE (1997) A decision-theoretic generalization of on-line learning and an application to boosting. J Comput Syst Sci 55 (1):119–139
Chang C-C, Lin C-J (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2(3):27
Quinlan JR (1996) Improved use of continuous attributes in C4. 5. J Artif Intell Res 4:77–90
Wilcoxon F (1945) Individual comparisons by ranking methods. Biometr Bull 1(6):80–83
Sheskin DJ (2003) Handbook of parametric and nonparametric statistical procedures. CRC Press, Boca Raton
Chembench repository. https://chembench.mml.unc.edu. Accessed 8 Aug 2018
Schattel V, Hinselmann G, Jahn A, Zell A, Laufer S (2011) Modeling and benchmark data set for the inhibition of c-Jun N-terminal kinase-3. J Chem Inf Model 51(3):670–679
Hammann F, Suenderhauf C, Huwyler Jr (2011) A binary ant colony optimization classifier for molecular activities. J Chem Inf Model 51(10):2690–2696
Mohr J, Jain B, Sutter A, Laak AT, Steger-Hartmann T, Heinrich N, Obermayer K (2010) A maximum common subgraph kernel method for predicting the chromosome aberration test. J Chem Inf Model 50(10):1821–1838
Russom CL, Williams CR, Stewart TW, Swank AE, Richard AM (2008) DSSTox EPA fathead minnow acute toxicity database (EPAFHM): SDF files and documentation, version: EPAFHM_v4b_617_15Feb2008, http://www.epa.gov/ncct/dsstox/sdf_epafhm.html. Accessed 8 Aug 2018
Fontaine F, Pastor M, Zamora I, Sanz F (2005) Anchor—grind: Filling the gap between standard 3d qsar and the grid-independent descriptors. J Med Chem 48(7):2687–2694
Helma C, Cramer T, Kramer S, De Raedt L (2004) Data mining and machine learning techniques for the identification of mutagenicity inducing substructures and structure activity relationships of noncongeneric compounds. J Chem Inf Comput Sci 44(4):1402–1411
Acknowledgements
This work was supported in part by Project TIN2015-66108-P of the Spanish Ministry of Science and Innovation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cerruela García, G., García-Pedrajas, N. Boosted feature selectors: a case study on prediction P-gp inhibitors and substrates. J Comput Aided Mol Des 32, 1273–1294 (2018). https://doi.org/10.1007/s10822-018-0171-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0171-5