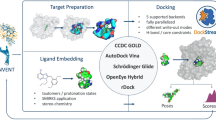
Abstract
Two of the major ongoing challenges in computational drug discovery are predicting the binding pose and affinity of a compound to a protein. The Drug Design Data Resource Grand Challenge 2 was developed to address these problems and to drive development of new methods. The challenge provided the 2D structures of compounds for which the organizers help blinded data in the form of 35 X-ray crystal structures and 102 binding affinity measurements and challenged participants to predict the binding pose and affinity of the compounds. We tested a number of pose prediction methods as part of the challenge; we found that docking methods that incorporate protein flexibility (Induced Fit Docking) outperformed methods that treated the protein as rigid. We also found that using binding pose metadynamics, a molecular dynamics based method, to score docked poses provided the best predictions of our methods with an average RMSD of 2.01 Å. We tested both structure-based (e.g. docking) and ligand-based methods (e.g. QSAR) in the affinity prediction portion of the competition. We found that our structure-based methods based on docking with Smina (Spearman ρ = 0.614), performed slightly better than our ligand-based methods (ρ = 0.543), and had equivalent performance with the other top methods in the competition. Despite the overall good performance of our methods in comparison to other participants in the challenge, there exists significant room for improvement especially in cases such as these where protein flexibility plays such a large role.







Similar content being viewed by others
References
Warren GL, Andrews CW, Capelli AM et al (2006) A critical assessment of docking programs and scoring functions. J Med Chem 49:5912–5931
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3:935–949
Damm-Ganamet KL, Smith RD, Dunbar JB Jr et al (2013) CSAR benchmark exercise 2011–2012: evaluation of results from docking and relative ranking of blinded congeneric series. J Chem Inf Model 53:1853–1870
Smith RD, Dunbar JB Jr, Ung PM et al (2011) CSAR benchmark exercise of 2010: combined evaluation across all submitted scoring functions. J Chem Inf Model 51:2115–2131
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Friesner RA, Banks JL, Murphy RB et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Oshiro CM, Kuntz ID, Dixon JS (1995) Flexible ligand docking using a genetic algorithm. J Comput Aided Mol Des 9:113–130
Jones G, Willett P (1995) Docking small-molecule ligands into active-sites. Curr Opin Biotechnol 6:652–656
Wang R, Lu Y, Wang S (2003) Comparative evaluation of 11 scoring functions for molecular docking. J Med Chem 46:2287–2303
Najmanovich R, Kuttner J, Sobolev V, Edelman M (2000) Side-chain flexibility in proteins upon ligand binding. Proteins Struct Funct Genet 39:261–268
Zavodszky MI (2005) Side-chain flexibility in protein-ligand binding: the minimal rotation hypothesis. Protein Sci 14:1104–1114
Brylinski M, Skolnick J (2008) What is the relationship between the global structures of apo and holo proteins? Proteins Struct Funct Genet 70:363–377
Boström J, Hogner A, Schmitt S (2006) Do structurally similar ligands bind in a similar fashion? J Med Chem 49:6716–6725
Sutherland JJ, Nandigam RK, Erickson JA, Vieth M (2007) Lessons in molecular recognition. 2. Assessing and improving cross-docking accuracy. J Chem Inf Model 47:2293–2302
Lin JH, Perryman AL, Schames JR, McCammon JA (2002) Computational drug design accommodating receptor flexibility: the relaxed complex scheme. J Am Chem Soc 124:5632–5633
Koes DR, Baumgartner MP, Camacho CJ (2013) Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J Chem Inf Model 53:1893–1904
Clark AJ, Tiwary P, Borrelli K et al (2016) Prediction of protein–ligand binding poses via a combination of induced fit docking and metadynamics simulations. J Chem Theory Comput 12:2990–2998
Sherman W, Day T, Jacobson MP et al (2006) Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem 49:534–553
Fletcher CM, Wagner G (1998) The interaction of eIF4E with 4E-BP1 is an induced fit to a completely disordered protein. Protein Sci 7:1639–1642
Prasad JC, Goldstone JV, Camacho CJ et al (2007) Ensemble modeling of substrate binding to cytochromes P450: analysis of catalytic differences between CYP1A orthologs. Biochemistry 46:2640–2654
Knegtel RM, Kuntz ID, Oshiro CM (1997) Molecular docking to ensembles of protein structures. J Mol Biol 266:424–440
Huang SY, Zou X (2007) Ensemble docking of multiple protein structures: considering protein structural variations in molecular docking. Proteins Struct Funct Genet 66:399–421
Luitz MP, Zacharias M (2014) Protein-ligand docking using Hamiltonian replica exchange simulations with soft core potentials. J Chem Inf Model 54:1669–1675
Colizzi F, Perozzo R, Scapozza L et al (2010) Single-molecule pulling simulations can discern active from inactive enzyme inhibitors. J Am Chem Soc 132:7361–7371
Di Nola A, Roccatano D, Berendsen HJ (1994) Molecular dynamics simulation of the docking of substrates to proteins. Proteins 19:174–182
Osguthorpe DJ, Sherman W, Hagler AT (2012) Exploring protein flexibility: incorporating structural ensembles from crystal structures and simulation into virtual screening protocols. J Phys Chem B 116:6952–6959
Wang K, Chodera JD, Yang Y, Shirts MR (2013) Identifying ligand binding sites and poses using GPU-accelerated Hamiltonian replica exchange molecular dynamics. J Comput Aided Mol Des 27:989–1007
Laio A, Gervasio FL, et al (2008) Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep Prog Phys 71:126601
Gervasio FL, Laio A, Parrinello M (2005) Flexible docking in solution using metadynamics. J Am Chem Soc 127:2600–2607
Sandak B, Wolfson HJ, Nussinov R (1998) Flexible docking allowing induced fit in proteins: insights from an open to closed conformational isomers. Proteins Struct Funct Genet 32:159–174
Claußen H, Buning C, Rarey M, Lengauer T (2001) FlexE: efficient molecular docking considering protein structure variations. J Mol Biol 308:377–395
Jorgensen WL (2004) The many roles of computation in drug discovery. Science 303:1813–1818
Jorgensen WL (2009) Efficient drug lead discovery and optimization. Acc Chem Res 42:724–733
Halgren TA, Murphy RB, Friesner RA et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750–1759
Jones G, Willett P, Glen RC et al (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748
Woo HJ, Roux B (2005) Calculation of absolute protein-ligand binding free energy from computer simulations. Proc Natl Acad Sci USA 102:6825–6830
Masetti M, Cavalli A, Recanatini M, Gervasio FL (2009) Exploring complex protein–ligand recognition mechanisms with coarse metadynamics. J Phys Chem B 113:4807–4816
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:27-28-38
De Vivo M, Masetti M, Bottegoni G, Cavalli A (2016) Role of molecular dynamics and related methods in drug discovery. J Med Chem 59:4035–4061
Durrant JD, McCammon JA (2011) Molecular dynamics simulations and drug discovery. BMC Biol 9:71
Cavalli A, Spitaleri A, Saladino G, Gervasio FL (2015) Investigating drug-target association and dissociation mechanisms using metadynamics-based algorithms. Acc Chem Res 48:277–285
Wolber G, Langer T (2005) LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model 45:160–169
Koes DR, Camacho CJ (2011) Pharmer: efficient and exact pharmacophore search. J Chem Inf Model 51:1307–1314
Wolber G, Seidel T, Bendix F, Langer T (2008) Molecule-pharmacophore superpositioning and pattern matching in computational drug design. Drug Discov Today 13:23–29
Cherkasov A, Muratov EN, Fourches D et al (2014) QSAR modeling: where have you been? Where are you going to? J Med Chem 57:4977–5010
Tropsha A (2010) Best practices for QSAR model development, validation, and exploitation. Mol Inform 29:476–488
Gathiaka S, Liu S, Chiu M et al (2016) D3R grand challenge 2015: evaluation of protein–ligand pose and affinity predictions. J Comput Aided Mol Des 30:651–668
Forman BM, Goode E, Chen J et al (1995) Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81:687–693
Jiao Y, Lu Y, Li X (2015) Farnesoid X receptor: a master regulator of hepatic triglyceride and glucose homeostasis. Acta Pharmacol Sin 36:44–50
Ye Z, Baumgartner MP, Wingert BM, Camacho CJ (2016) Optimal strategies for virtual screening of induced-fit and flexible target in the 2015 D3R Grand Challenge. J Comput Aided Mol Des 30:695–706
Baumgartner MP, Camacho CJ (2016) Choosing the optimal rigid receptor for docking and scoring in the CSAR 2013/2014 experiment. J Chem Inf Model 56:1004–1012
Berman HM (2000) The protein data bank. Nucleic Acids Res 28:235–242
Rose PW, Prlić A, Altunkaya A et al (2016) The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res 45(D1):D271–D281
Small-Molecule Drug Discovery Suite 2016-3 (2016) Schrödinger, LLC, New York, NY
Madhavi Sastry G, Adzhigirey M, Day T et al (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234
Jacobson MP, Pincus DL, Rapp CS et al (2004) A hierarchical approach to all-atom protein loop prediction. Proteins Struct Funct Genet 55:351–367
Jacobson MP, Friesner RA, Xiang Z, Honig B (2002) On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol 320:597–608
Harder E, Damm W, Maple J et al (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12:281–296
Chemical Computing Group ULC (2017) Molecular operating environment (MOE), 2013.08. Montreal, QC, Canada
Durant JL, Leland BA, Henry DR, Nourse JG (2002) Reoptimization of MDL keys for use in drug discovery. J Chem Inf Comput Sci 42:1273–1280
Gao C, Thorsteinson N, Watson I et al (2015) Knowledge-based strategy to improve ligand pose prediction accuracy for lead optimization. J Chem Inf Model 55:1460–1468
POSIT 3.1.0.5: OpenEye Scientific Software, Santa Fe, NM. http://www.eyesopen.com
Watts KS, Dalal P, Murphy RB et al (2010) ConfGen: a conformational search method for efficient generation of bioactive conformers. J Chem Inf Model 50:534–546
Forge, 10.4.0, Cresset, Litlington, Cambridgeshire, UK. http://www.cresset-group.com/forge/
Richter HGF, Benson GM, Blum D et al (2011) Discovery of novel and orally active FXR agonists for the potential treatment of dyslipidemia & diabetes. Bioorg Med Chem Lett 21:191–194
Richter HGF, Benson GM, Bleicher KH et al (2011) Optimization of a novel class of benzimidazole-based farnesoid X receptor (FXR) agonists to improve physicochemical and ADME properties. Bioorg Med Chem Lett 21:1134–1140
Akwabi-Ameyaw A, Bass JY, Caldwell RD et al (2008) Conformationally constrained farnesoid X receptor (FXR) agonists: naphthoic acid-based analogs of GW 4064. Bioorg Med Chem Lett 18:4339–4343
Sherman W, Beard HS, Farid R (2006) Use of an induced fit receptor structure in virtual screening. Chem Biol Drug Des 67:83–84
Farid R, Day T, Friesner RA, Pearlstein RA (2006) New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg Med Chem 14:3160–3173
Dolgikh E, Watson IA, Desai PV et al (2016) QSAR model of unbound brain-to-plasma partition coefficient, Kp,uu,brain: incorporating P-glycoprotein efflux as a variable. J Chem Inf Model 56:2225–2233
PyMOL: The PyMOL Molecular Graphics System, Version 1.8.6.0 Schrödinger, LLC
Acknowledgements
The research leading to these results has received funding from the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement No. 612347. The authors wish to acknowledge Schrödinger, LLC for providing temporary software licenses for this work. The authors would like to thank Davide Branduardi (Schrödinger) for helpful discussion and advice. The authors thank Ian Watson (Eli Lilly) for his assistance and advice with the ligand-based scoring methods and Lewis Vidler (Eli Lilly) for his critical feedback and discussion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Baumgartner, M.P., Evans, D.A. Lessons learned in induced fit docking and metadynamics in the Drug Design Data Resource Grand Challenge 2. J Comput Aided Mol Des 32, 45–58 (2018). https://doi.org/10.1007/s10822-017-0081-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-017-0081-y