Abstract
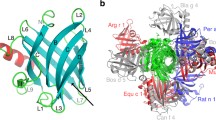
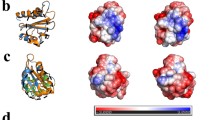
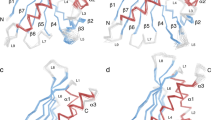
Alt a 1 is a highly allergenic protein from Alternaria fungi responsible for several respiratory diseases. Its crystal structure revealed a unique β-barrel fold that defines a new family exclusive to fungi and forms a symmetrical dimer in a butterfly-like shape as well as tetramers. Its biological function is as yet unknown but its localization in cell wall of Alternaria spores and its interactions in the onset of allergy reactions point to a function to transport ligands. However, at odds with binding features in β-barrel proteins, monomeric Alt a 1 seems unable to harbor ligands because the barrel is too narrow. Tetrameric Alt a 1 is able to bind the flavonoid quercetin, yet the stability of the aggregate and the own ligand binding are pH-dependent. At pH 6.5, which Alt a 1 would meet when secreted by spores in bronchial epithelium, tetramer-quercetin complex is stable. At pH 5.5, which Alt a 1 would meet in apoplast when infecting plants, the complex breaks down. By means of a combined computational study that includes docking calculations, empirical pKa estimates, Poisson–Boltzmann electrostatic potentials, and Molecular Dynamics simulations, we identified a putative binding site at the dimeric interface between subunits in tetramer. We propose an explanation on the pH-dependence of both oligomerization states and protein–ligand affinity of Alt a 1 in terms of electrostatic variations associated to distinct protonation states at different pHs. The uniqueness of this singular protein can thus be tracked in the combination of all these features.








Similar content being viewed by others
Abbreviations
- 3D:
-
Three-dimensional
- APBS:
-
Adaptive Poisson–Boltzmann solver
- DPBA:
-
Diphenylboric acid 2-aminoethyl ester
- MD:
-
Molecular dynamics
- PB:
-
Poisson–Boltzmann
- PDB:
-
Protein data bank
- PISA:
-
Proteins, interfaces, structures, and assemblies
- PTGL:
-
Protein topology graph library
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
References
Delfino RJ, Zeiger RS, Seltzer JM, Street DH, Matteucci RM, Anderson PR et al (1997) The effect of outdoor fungal spore concentrations on daily asthma severity. Environ Health Perspect 105:622–635
Bush RK, Portnoy JM (2001) The role and abatement of fungal allergens in allergic diseases. J Allergy Clin Immunol 107:S430–S440
Anderson M, Downs S, Mitakatis T, Leuppi J, Marks G (2003) Natural exposure to Alternaria spores induces allergic rhinitis symptoms in sensitized children. Pediatr Allergy Immunol 14:100–105
Bush RK, Prochnau JJ (2004) Alternaria-induced asthma. J Allergy Clin Immunol 113:227–234
Knutsen AP, Bush RK, Demain JG, Denning DW, Dixit A, Fairs A et al (2012) Fungi and allergic lower respiratory tract diseases. J Allergy Clin Immunol 129:280–289
Salo PM, Arbes SJ, Sever M, Jaramillo R, Cohn RD, London SJ, Zeldin DC (2006) Exposure to Alternaria alternata in US homes is associated with asthma symptoms. J Allergy Clin Immunol 118:892–898
Feo Brito F, Alonso AM, Carnes J, Martin-Martin R, Fernandez-Caldas E, Galindo PA et al (2012) Correlation between Alt a 1 levels and clinical symptoms in Alternaria alternata-monosensitized patients. J Invest Allergol Clin Immunol 22:154–159
Deards MJ, Montague AE (1991) Purification and characterization of a major allergen of Alternaria alternata. Mol Immunol 28:409–415
De Vouge MW, Thaker AJ, Curran IH, Zhang L, Muradia G, Rode H et al (1996) Isolation and expression of a cDNA clone encoding an Alternaria alternata Alt a 1 subunit. Int Arch Allergy Immunol 111:385–395
Vailes LD, Perzanowski MS, Wheatley LM, Platt-Mills TA, Chapman MD (2001) IgE and IgG antibody responses to recombinant Alt a 1 as a marker of sensitization to Alternaria in asthma and atopic dermatitis. Clin Exp Allergy 31:1891–1895
Asturias JA, Ibarrola I, Ferrer A, Andreu C, Lopez-Pascual E, Quiralte J et al (2005) Diagnosis of Alternaria alternata sensitization with natural and recombinant Alt a 1 allergens. J Allergy Clin Immunol 115:1210–1217
Twaroch TE, Focke M, Fleischmann K, Balic N, Lupinek K, Blatt K et al (2012) Carrier-bound Alt a 1 peptides without allergenic activity for vaccination against Alternaria alternata allergy. Clin Exp Allergy 42:966–975
Kurup VP, Vijay HM, Kumar V, Castillo L, Elms N (2003) IgE binding synthetic peptides of Alt a 1, a major allergen of Alternaria alternata. Peptides 24:179–185
Chrusczc M, Chapman MD, Osinski T, Solberg R, Demas M, Porebski PJ et al (2012) Alternaria alternata allergen Alt a 1: a unique β-barrel protein dimer found exclusively in fungi. J Allergy Clin Immunol 130:241–247
Rouvinen J, Janis J, Laukhanen ML, Jylha S, Niemi M, Paivinen T et al (2010) Transient dimers of allergens. PLoS One 5:e9037/1-9
Wagner GE, Gutfreund S, Fauland K, Keller W, Valenta R, Zangger K (2014) Backbone resonance assignment of Alt a 1, a unique β-barrel protein and the major allergen of Alternaria alternata. Biomol NMR Assign 8:229–231
Twaroch TE, Arcalis E, Sterflinger K, Stoger E, Swoboda I, Valenta R (2012) Predominant localization of the major Alternaria allergen Alt a 1 in the cell wall of airborne spores. J Allergy Clin Immunol 129:1148–1149
Mitakakis TZ, Barnes C, Tovey ER (2011) Spore germination increases allergen release from Alternaria. J Allergy Clin Immunol 107:388–390
Garrido-Arandia M, Gómez-Casado C, Díaz-Perales A, Pacios LF (2014) Molecular dynamics of major allergens from Alternaria, birch pollen and peach. Mol Inf 33:682–694
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Touw WG, Baakman C, Black J, Te Beek TAH, Krieger E, Robbie P et al (2015) A series of PDB related databases for everyday needs. Nucleic Acids Res 43:D364–D368
Laskowski RA (2001) PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res 29:221–222
Laskowski RA (2014) PDBsum additions. Nucleic Acids Res 37:D355–D359
May P, Barthel S, Koch I (2004) Protein topology graph library. Bioinformatics 20:3277–3279
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797
Krissinel E (2010) Crystal contacts as nature’s docking solutions. J Comput Chem 31:133–143
The PyMOL Molecular Graphics System, Version 1.7.6.4 Schrödinger, LLC
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Siegel D, Troyanov S, Noack J, Emmerling F, Nehls I (2010) Alternariol. Acta Crystallogr Sect E 66:1366/1-7
Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theor Comput 11:3696–3713
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25:247–260
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 16:2785–2791
Trott O, Olson AJ (2009) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH (2011) PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theor Comput 7:525–537
Sondergaard CR, Olsson MHM, Rostkowski M, Jensen JH (2011) Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J Chem Theor Comput 7:2284–2295
Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup, execution, and analysis of Poisson-Boltzmann electrostatics calculations. Nucl Acids Res 32:W665–W667
Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucl Acids Res 35:W522–W525
MacKerell A, Bashford D, Bellott M, Dunbrack RL, Evanseck J, Field MJ et al (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616
Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98:10037–10041
Zoete V, Cuendet MA, Grosdidier A, Michielin O (2011) SwissParam, a fast force field generation tool for small organic molecules. J Comput Chem 32:2359–2368
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E et al (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graph 14:33–38
Fischer F, Wissicombe JH (2006) Mechanism of acid and base secretion by the airway epithelium. J Membr Biol 211:139–150
Almeida DPF, Huber DJ (1999) Apoplastic pH and inorganic ion levels in tomato fruit: a potential means for regulation of cell wall metabolism during ripening. Physiol Plant 105:506–512
Cianci M, Folli C, Zonta F, Florio P, Berni R, Zanotti G (2015) Structural evidence for asymmetric ligand binding to transthyretin. Acta Crystallogr Sect D 71:1582–1592
Pasquato N, Berni R, Folli C, Alfieri B, Cendron L, Zanotti G (2007) Acidic pH-induced conformational change in amyloidogenic mutant transthyretin. J Mol Biol 366:711–719
Barkai-Golan R (2008) Alternaria mycotoxins. In: Barkai-Golan R, Nachman P (eds) Mycotoxins in fruits and vegetables. Academic Press, San Diego, pp 185–203
European Food Safety Authority (2011) Scientific opinion on the risks for animal and public health related to the presence of Alternaria toxins in feed and food. EFSA J 9(2407):1–97
Garrido-Arandia M, Gómez-Casado C, Díaz-Perales A, Pacios LF (2014) Distortion from planarity in arenes produced by internal rotation of one single hydroxyl hydrogen: the case of alternariol. J Mol Graph Model 53:140–147
Acknowledgments
All Molecular Dynamics calculations were carried out on the Magerit supercomputer of Universidad Politécnica de Madrid. The authors acknowledge the computer resources and technical assistance provided by the Centro de Supercomputación y Visualización de Madrid (CeSViMa). This work was supported by the Spanish Ministerio de Ciencia e Innovación MINECO (Grant BIO2013-41403-R) and Instituto de Salud Carlos III, Spain, RETICS 2007 (RD12/0013/14). CGC was supported by the FPI programme from Spanish Government (MICINN/MINECO, Grant BES-2010-034628).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Garrido-Arandia, M., Bretones, J., Gómez-Casado, C. et al. Computational study of pH-dependent oligomerization and ligand binding in Alt a 1, a highly allergenic protein with a unique fold. J Comput Aided Mol Des 30, 365–379 (2016). https://doi.org/10.1007/s10822-016-9911-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-016-9911-6