Abstract
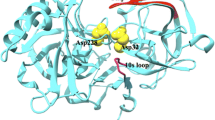
The identification of hot spots, i.e., binding regions that contribute substantially to the free energy of ligand binding, is a critical step for structure-based drug design. Here we present the application of two fragment-based methods to the detection of hot spots for DJ-1 and glucocerebrosidase (GCase), targets for the development of therapeutics for Parkinson’s and Gaucher’s diseases, respectively. While the structures of these two proteins are known, binding information is lacking. In this study we employ the experimental multiple solvent crystal structures (MSCS) method and computational fragment mapping (FTMap) to identify regions suitable for the development of pharmacological chaperones for DJ-1 and GCase. Comparison of data derived via MSCS and FTMap also shows that FTMap, a computational method for the identification of fragment binding hot spots, is an accurate and robust alternative to the performance of expensive and difficult crystallographic experiments.








Similar content being viewed by others
References
Bembenek SD, Tounge BA, Reynolds CH (2009) Drug Discov Today 14:278–283. doi:10.1016/j.drudis.2008.11.007
Ciulli A, Williams G et al (2006) J Med Chem 49:4992–5000. doi:10.1021/jm060490r
Parkinson J (1817) An essay on the shaking palsy. Whitingham and Rowland, London
Jankovic J (2008) J Neurol Neurosurg Psychiatry 79:368–376. doi:10.1136/jnnp.2007.131045
Bonifati V (2007) Parkinsonism Relat Disord 13(Suppl 3):S233–S241. doi:10.1016/S1353-8020(08)70008-7
da Costa CA (2007) Curr Mol Med 7:650–657. doi:10.2174/156652407782564426
Biskup S, Gerlach M et al (2008) J Neurol 255(Suppl 5):8–17. doi:10.1007/s00415-008-5005-2
Wilson MA, Collins JL et al (2003) Proc Natl Acad Sci USA 100:9256–9261. doi:10.1073/pnas.1133288100
Moore DJ, Zhang L et al (2003) J Neurochem 87:1558–1567
Miller DW, Ahmad R et al (2003) J Biol Chem 278:36588–36595. doi:10.1074/jbc.M304272200
Rohrbach M, Clarke JT (2007) Drugs 67:2697–2716. doi:10.2165/00003495-200767180-00005
Liou B, Kazimierczuk A et al (2006) J Biol Chem 281:4242–4253. doi:10.1074/jbc.M511110200
Schmitz M, Alfalah M et al (2005) Int J Biochem Cell Biol 37:2310–2320. doi:10.1016/j.biocel.2005.05.008
Grace ME, Newman KM et al (1994) J Biol Chem 269:2283–2291
Yu Z, Sawkar AR, Kelly JW (2007) FEBS 274:4944–4950. doi:10.1111/j.1742-4658.2007.06042.x
Steet RA, Chung S et al (2006) Proc Natl Acad Sci USA 103:13813–13818. doi:10.1073/pnas.0605928103
Sawkar AR, Zimmer KD et al (2006) ACS Chem Biol 1:235–251. doi:10.1021/cb600187q
Compain P, Martin OR et al (2006) ChemBioChem 7:1356–1359. doi:10.1002/cbic.200600217
Sawkar AR, D’Haeze W, Kelley JW (2006) Cell Mol Life Sci 63:1179–1192. doi:10.1007/s00018-005-5437-0
Sawkar AR, Adamski-Werner SL et al (2005) Chem Biol 12:1235–1244. doi:10.1016/j.chembiol.2005.09.007
Sawkar AR, Cheng WC et al (2002) Proc Natl Acad Sci USA 99:15428–15433. doi:10.1073/pnas.192582899
Lieberman RL, Wustman BA et al (2007) Nat Chem Biol 3:101–107. doi:10.1038/nchembio850
Allen KN, Bellamacina CR et al (1996) J Phys Chem 100:2605–2611. doi:10.1021/jp952516o
Mattos C, Bellamacina CR et al (2006) J Mol Biol 357:1471–1482. doi:10.1016/j.jmb.2006.01.039
Mattos C, Ringe D (1996) Nat Biotechnol 14:595–599. doi:10.1038/nbt0596-595
Brenke R, Kozakov D et al (2009) Bioinformatics 25:621–627. doi:10.1093/bioinformatics/btp036
Dennis S, Kortvelyesi T, Vajda S (2002) Proc Natl Acad Sci USA 99:4290–4295. doi:10.1073/pnas.062398499
Kortvelyesi T, Dennis S et al (2003) Proteins 51:340–351. doi:10.1002/prot.10287
Silberstein M, Dennis S et al (2003) J Mol Biol 332:1095–1113. doi:10.1016/j.jmb.2003.08.019
Goodford PJ (1985) J Med Chem 28:849–857. doi:10.1021/jm00145a002
Stultz CM, Karplus M (1999) Proteins 37:512–529. doi:10.1002/(SICI)1097-0134(19991201)37:4<512::AID-PROT3>3.0.CO;2-O
Landon MR, Lancia DR et al (2007) J Med Chem 50:1231–1240. doi:10.1021/jm061134b
Otwinowski Z, Minor W (1997) Methods Enzymol Macromol Crystallogr Pt A 276:307–326
Vagin A, Teplyakov A (1997) J Appl Cryst 30:1022–1025. doi:10.1107/S0021889897006766
Murshudov GN, Vagin AA, Dodson EJ (1997) Acta Crystallogr D Biol Crystallogr 53:240–255. doi:10.1107/S0907444996012255
Collaborative Computational Project, Number 4 (1994) Acta Crystallogr D Biol Crystallogr 50:760–763. doi:10.1107/S0907444994003112
Emsley P, Cowtan K (2004) Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi:10.1107/S0907444904019158
Kozakov D, Brenke R et al (2006) Proteins 65:392–406. doi:10.1002/prot.21117
Schaefer M, Karplus M (1996) J Phys Chem 100:1578–1599. doi:10.1021/jp9521621
Brooks BR, Bruccoleri RE et al (1983) J Comput Chem 4:187–217. doi:10.1002/jcc.540040211
Kosakov D, Clodfelter K et al (2005) Biophys J 89:867–875. doi:10.1529/biophysj.104.058768
Ruvinsky AM, Kozintsev AV (2006) Proteins 62:202–208. doi:10.1002/prot.20673
Blackinton R, Lakshminarasimhan M et al (2009) J Biol Chem 284:6476–6485. doi:10.1074/jbc.M806599200
Delano WL (2008) The PyMol molecular graphics system. Delano Scientific, Palo Alto
Brumshtein B, Wormald MR et al (2006) Acta Crystallogr D Biol Crystallogr 62:1458–1465. doi:10.1107/S0907444906038303
Kacher Y, Brumshtein B et al (2008) Biol Chem 389:1361–1369. doi:10.1515/BC.2008.163
Salvioli R, Tatti M et al (2005) Biochem J 390:95–103. doi:10.1042/BJ20050325
Reczek D, Schwake M et al (2007) Cell 131:770–783. doi:10.1016/j.cell.2007.10.018
de Alba E, Weiler S, Tjandra N (2003) Biochemistry 42:14729–14740. doi:10.1021/bi0301338
Hawkins CA, de Alba E, Tjandra N (2005) J Mol Biol 346:1381–1392. doi:10.1016/j.jmb.2004.12.045
John M, Wendeler M et al (2006) Biochemistry 45:5206–5216. doi:10.1021/bi051944+
Acknowledgments
M. R. L was supported by grant F32NS061415 from the National Institute of Neurological Disorders and Stroke (NINDS). Research performed in the laboratory of S. V. was supported by grant GM064700 from the National Institutes of Health (NIH). R. L. L. was supported by fellowship F32AG027647 from the National Institutes of Health. G. A. P. is a Duvoisin fellow of the American Parkinson’s Disease Association. G. A. P. and D. R. are recipients of an award from the McKnight Endowment Fund for Neuroscience. Parkinson’s Disease work at Brandeis University was initiated with generous support from the Ellison Medical Foundation. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL) and the Advanced Photo Source (APS), national user facilities operated on behalf of the US Department of Energy, Office of Basic Energy Sciences. Work performed at FM/CA-CAT at APS has been funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). We would also like to thank Amicus Therapeutics for their generous support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Landon, M.R., Lieberman, R.L., Hoang, Q.Q. et al. Detection of ligand binding hot spots on protein surfaces via fragment-based methods: application to DJ-1 and glucocerebrosidase. J Comput Aided Mol Des 23, 491–500 (2009). https://doi.org/10.1007/s10822-009-9283-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-009-9283-2