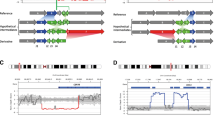
Abstract
Purpose
To report genetic characteristics and associated risk of chromosomal breaks due to chromosomal rearrangements in large samples.
Methods
MicroSeq, a technique that combines chromosome microdissection and next-generation sequencing, was used to identify chromosomal breakpoints. Long-range PCR and Sanger sequencing were used to precisely characterize 100 breakpoints in 50 ABCR carriers.
Results
In addition to the recurrent regions of balanced rearrangement breaks in 8q24.13, 11q11.23, and 22q11.21 that had been documented, we have discovered a 10-Mb region of 12q24.13-q24.3 that could potentially be a sparse region of balanced rearrangement breaks. We found that 898 breakpoints caused gene disruption and a total of 188 breakpoints interrupted genes recorded in OMIM. The percentage of breakpoints that disrupted autosomal dominant genes recorded in OMIM was 25.53% (48/188). Fifty-four of the precisely characterized breakpoints had 1–8-bp microhomologous sequences.
Conclusion
Our findings provide a reference for the evaluation of the pathogenicity of mutations in related genes that cause protein truncation in clinical practice. According to the characteristics of breakpoints, non-homologous end joining and microhomology-mediated break-induced replication may be the main mechanism for ABCRs formation.




Similar content being viewed by others
References
Jacobs PA, et al. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J Med Genet. 1992;29:103–8. https://doi.org/10.1136/jmg.29.2.103.
Giardino D, et al. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenat Diagn. 2009;29:257–65. https://doi.org/10.1002/pd.2215.
Halgren C, et al. Risks and recommendations in prenatally detected de novo balanced chromosomal rearrangements from assessment of long-term outcomes. Am J Hum Genet. 2018;102:1090–103. https://doi.org/10.1016/j.ajhg.2018.04.005.
David D, et al. Comprehensive clinically oriented workflow for nucleotide level resolution and interpretation in prenatal diagnosis of de novo apparently balanced chromosomal translocations in their genomic landscape. Hum Genet. 2020;139:531–43. https://doi.org/10.1007/s00439-020-02121-x.
Gijsbers AC, et al. Whole genome paired-end sequencing elucidates functional and phenotypic consequences of balanced chromosomal rearrangement in patients with developmental disorders. Eur J Med Genet. 2019;56:526–35. https://doi.org/10.1136/jmedgenet-2018-105778.
Schluth-Bolard C, et al. Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: array CGH study of 47 unrelated cases. Eur J Med Genet. 2009;52(5):291–6. https://doi.org/10.1016/j.ejmg.2009.05.011.
Feenstra I, et al. Balanced into array: genome-wide array analysis in 54 patients with an apparently balanced de novo chromosome rearrangement and a meta-analysis. Eur J Hum Genet. 2011;19:1152–60. https://doi.org/10.1038/ejhg.2011.120.
Dong Z, et al. Genome sequencing explores complexity of chromosomal abnormalities in recurrent miscarriage. Am J Hum Genet. 2019;105:1102–11. https://doi.org/10.1016/j.ajhg.2019.10.003.
Miao ZY, et al. Cytogenetic analysis of 2959 couples with spontaneous abortion and detailed analysis of rare karyotypes. J Genet. 2022;101:10.
Chau MHK, et al. Investigation of the genetic etiology in male infertility with apparently balanced chromosomal structural rearrangements by genome sequencing. Asian J Androl. 2022;24(3):248–54. https://doi.org/10.4103/aja2021106.
Carvalho CM, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17:224–38. https://doi.org/10.1038/nrg.2015.25.
Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. https://doi.org/10.1074/jbc.R700039200.
Mitelman F, et al. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. https://doi.org/10.1038/nrc2091.
Mitelman F, et al. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosomes Cancer. 2005;43:350–66. https://doi.org/10.1002/gcc.20212.
Schnause AC, et al. Marfan syndrome caused by disruption of the FBN1 gene due to a reciprocal chromosome translocation. Genes (Basel). 2021;12:1836. https://doi.org/10.3390/genes12111836.
Siffroi JP, et al. Assisted reproductive technology and complex chromosomal rearrangements: the limits of ICSI. Mol Hum Reprod. 1997;3:847–51. https://doi.org/10.1093/molehr/3.10.847.
Miny P, Schloo R. Ist Sterilität eine Erblast? [Is sterility a genetic burden?]. Ther Umsch. 1999;56:265–70. https://doi.org/10.1024/0040-5930.56.5.265.
Neri G, et al. Reproductive risks for translocation carriers: cytogenetic study and analysis of pregnancy outcome in 58 families. Am J Med Genet. 1983;16:535–61. https://doi.org/10.1002/ajmg.1320160412.
Park JM, et al. An autosomal dominant ERLIN2 mutation leads to a pure HSP phenotype distinct from the autosomal recessive ERLIN2 mutations (SPG18). Sci Rep. 2020;10:3295. https://doi.org/10.1038/s41598-020-60374-y.
Cheon CK, et al. Autosomal dominant transmission of complicated hereditary spastic paraplegia due to a dominant negative mutation of KIF1A, SPG30 gene. Sci Rep. 2017;7:12527. https://doi.org/10.1038/s41598-017-12999-9.
Couch FJ, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol. 2015;33:304–11. https://doi.org/10.1200/JCO.2014.57.1414.
Couch FJ, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol Sep. 2017;3(9):1190–6. https://doi.org/10.1001/jamaoncol.2017.0424.
Kilpivaara O, et al. CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer. 2004;111:43–547. https://doi.org/10.1002/ijc.20299.
Lieber MR, et al. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–33. https://doi.org/10.1038/cr.2007.108.
Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–22. https://doi.org/10.1016/s0168-9525(98)01555-8.
Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. https://doi.org/10.1016/s0168-9525(02)02592-1.
Lee JA, et al. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235–47. https://doi.org/10.1016/j.cell.2007.11.037.
Hastings PJ, et al. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5(1):e1000327. https://doi.org/10.1371/journal.pgen.1000327.
Burssed B, et al. Mechanisms of structural chromosomal rearrangement formation. Mol Cytogenet. 2009;15:23. https://doi.org/10.1186/s13039-022-00600-6.
Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–71. https://doi.org/10.1146/annurev-genet-110410-132435.
Nilsson D, et al. Whole-genome sequencing of cytogenetically balanced chromosome translocations identifies potentially pathological gene disruptions and highlights the importance of microhomology in the mechanism of formation. Hum Mutat. 2017;38:180–92. https://doi.org/10.1002/humu.23146.
Hu L, et al. Reciprocal translocation carrier diagnosis in preimplantation human embryos. EBioMedicine. 2016;14:139–47. https://doi.org/10.1016/j.ebiom.2016.11.007.
Levy B, Stosic M. Traditional prenatal diagnosis: past to present. Methods Mol Biol. 2019;1885:3–22. https://doi.org/10.1007/978-1-4939-8889-1_1.
D'Alton ME, DeCherney AH. Prenatal diagnosis. N Engl J Med. 1993;328:114–20. https://doi.org/10.1056/NEJM199301143280208.
Sullivan-Pyke C, Dokras A. Preimplantation genetic screening and preimplantation genetic diagnosis. Obstet Gynecol Clin North Am. 2018;45:113–25. https://doi.org/10.1016/j.ogc.2017.10.009.
Simpson JL, et al. Overview of preimplantation genetic diagnosis (PGD): historical perspective and future direction. Methods Mol Biol. 2019;1885:23–43. https://doi.org/10.1007/978-1-4939-8889-1_2.
Tan YQ, et al. Single-nucleotide polymorphism microarray-based preimplantation genetic diagnosis is likely to improve the clinical outcome for translocation carriers. Hum Reprod. 2013;28:2581–92. https://doi.org/10.1093/humrep/det271.
Trunca C, et al. Reproductive risk estimation calculator for balanced translocation carriers. Curr Protoc. 2022;2:e633. https://doi.org/10.1002/cpz1.633.
Redin C, et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet. 2017;49(1):36–45. https://doi.org/10.1038/ng.3720.
Wolff M, et al. Phenotypic spectrum and genetics of SCN2A-related disorders, treatment options, and outcomes in epilepsy and beyond. Epilepsia. 2019;60(Suppl 3):S59–67. https://doi.org/10.1111/epi.14935.
Kayvanpour E, et al. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin Res Cardiol. 2017;106:127–39. https://doi.org/10.1007/s00392-016-1033-6.
Ghavi-Helm Y. Functional consequences of chromosomal rearrangements on gene expression: not so deleterious after all? J Mol Biol. 2022;432:665–75. https://doi.org/10.1016/j.jmb.2019.09.010.
Lupiáñez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25. https://doi.org/10.1016/j.cell.2015.04.004.
Dong Z, et al. Identification of balanced chromosomal rearrangements previously unknown among participants in the 1000 Genomes Project: implications for interpretation of structural variation in genomes and the future of clinical cytogenetics. Genet Med. 2018;20:697–707. https://doi.org/10.1038/gim.2017.170.
Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet. 2004;13:R57–64. https://doi.org/10.1093/hmg/ddh073.
Chiang C, et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet. 2012;44:390–S1. https://doi.org/10.1038/ng.2202.
Conrad DF, et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–91. https://doi.org/10.1038/ng.564.
Dong Z, et al. Development of coupling controlled polymerizations by adapter-ligation in mate-pair sequencing for detection of various genomic variants in one single assay. DNA research: an international journal for rapid publication of reports on genes and genomes. DNA Res. 2019;26(4):313–25. https://doi.org/10.1093/dnares/dsz011.
Dong Z, et al. Deciphering the complexity of simple chromosomal insertions by genome sequencing. Hum Genet. 2021;140(2):361–80. https://doi.org/10.1007/s00439-020-02210-x.
Acknowledgements
We thank the patients and family members for their participation. Thanks to the National Natural Science Foundation of China for the strong support to this project.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81873478).
Author information
Authors and Affiliations
Contributions
Conceptualization: DC, YT, LH, GL; methodology: DC, GL, YT, LH; formal analysis and investigation: DC, YX, KL, ML; writing—original draft preparation: DC, YX; writing—review and editing: YX, LH; visualization: YX; funding acquisition: LH; resources: YT, GL, LH; supervision: LH.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by Reproductive and Genetic Hospital of CITIC Xiangya Ethics Committee and informed consent was obtained from the patients. The ethical approval number is LL-SC-2016-002.
Consent for publication
All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, Y., Cheng, D., Luo, K. et al. Evaluation of genetic risk of apparently balanced chromosomal rearrangement carriers by breakpoint characterization. J Assist Reprod Genet 41, 147–159 (2024). https://doi.org/10.1007/s10815-023-02986-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-023-02986-7