Abstract
Purpose
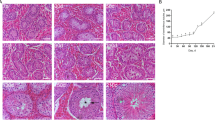
Rapid and easy detection of spermatogonial stem/progenitor cells (SSPCs) is crucial for clinicians dealing with male infertility caused by prepubertal testicular damage. Deep learning (DL) methods may offer visual tools for tracking SSPCs on testicular strips of prepubertal animal models. The purpose of this study is to detect and count the seminiferous tubules and SSPCs in newborn mouse testis sections using a DL method.
Methods
Testicular sections of the C57BL/6-type newborn mice were obtained and enumerated. Odd-numbered sections were stained with hematoxylin and eosin (H&E), and even-numbered sections were immune labeled (IL) with SSPC specific marker, SALL4. Seminiferous tubule and SSPC datasets were created using odd-numbered sections. SALL4-labeled sections were used as positive control. The YOLO object detection model based on DL was used to detect seminiferous tubules and stem cells.
Results
Test scores of the DL model in seminiferous tubules were obtained as 0.98 mAP, 0.93 precision, 0.96 recall, and 0.94 f1-score. The SSPC test scores were obtained as 0.88 mAP, 0.80 precision, 0.93 recall, and 0.82 f1-score.
Conclusion
Seminiferous tubules and SSPCs on prepubertal testicles were detected with a high sensitivity by preventing human-induced errors. Thus, the first step was taken for a system that automates the detection and counting process of these cells in the infertility clinic.





Similar content being viewed by others
Code availability
References
Skaznik-Wikiel ME, Gilbert SB, Meacham RB, Kondapalli LA. Fertility preservation options for men and women with cancer. Rev Urol. 2015;17(4):211–9. https://doi.org/10.3909/riu0666.
Wasilewski-Masker K, Seidel KD, Leisenring W, Mertens AC, Shnorhavorian M, Ritenour CW, Stovall M, Green DM, Sklar CA, Armstrong GT, Robison LL, Meacham LR. Male infertility in long-term survivors of pediatric cancer: a report from the childhood cancer survivor study. J Cancer Surviv. 2014;8(3):437–47. https://doi.org/10.1007/s11764-014-0354-6.
Levine JM. Preserving fertility in children and adolescents with cancer. Children. 2014;1(2):166–85. https://doi.org/10.3390/children1020166.
Önen S, Köse S, Yersal N, Korkusuz P. Mesenchymal stem cells promote spermatogonial stem/progenitor cell pool and spermatogenesis in neonatal mice in vitro. Sci Rep. 2022;12(1):11494. https://doi.org/10.1038/s41598-022-15358-5.
Yersal N, Köse S, Horzum U, Özkavukcu S, Orwig KE, Korkusuz P. Leptin promotes proliferation of neonatal mouse stem/progenitor spermatogonia. J Assist Reprod Genet. 2020;37(11):2825–38. https://doi.org/10.1007/s10815-020-01929-w.
Kubota H, Brinster RL. Spermatogonial stem cells. Biol Reprod. 2018;99(1):52–74. https://doi.org/10.1093/biolre/ioy077.
Lovelace DL, Gao Z, Mutoji K, Song YC, Ruan J, Hermann BP. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development. 2016;143(11):1893–906. https://doi.org/10.1242/dev.132761.
Korkusuz P, Köse S, Yersal N, Önen S. Magnetic-based cell isolation technique for the selection of stem cells. Methods Mol Biol. 2019;1879:153–63. https://doi.org/10.1007/7651_2018_151.
Köse S, Yersal N, Önen S, Korkusuz P. Comparison of hematopoietic and spermatogonial stem cell niches from the regenerative medicine aspect. Adv Exp Med Biol. 2018;1107:15–40. https://doi.org/10.1007/5584_2018_217.
Amisha, Malik P, Pathania M, Rathaur V. Overview of artificial intelligence in medicine. J Family Med Primary Care. 2019;8(7):2328–31. https://doi.org/10.4103/jfmpc.jfmpc_440_19.
Louis CM, Erwin A, Handayani N, Polim AA, Boediono A, Sini I. Review of computer vision application in in vitro fertilization: the application of deep learning-based computer vision technology in the world of IVF. J Assist Reprod Genet. 2021. https://doi.org/10.1007/s10815-021-02123-2.
Chan YK, Chen YF, Pham T, Chang WD, Hsieh MY. Artificial intelligence in medical applications. J Healthc Eng. 2018:2. https://doi.org/10.1155/2018/4827875.
Xu J, Lu H, Li H, Wang X, Madabhushi A, Xu Y. Histopathological image analysis on mouse testes for automated staging of mouse seminiferous tubule. Cham: Springer International Publishing; 2019. https://doi.org/10.1007/978-3-030-23937-4_14.
Xu J, Lu H, Li H, Yan C, Wang X, Zang M, Rooij DG, Madabhushi A, Xu EY. Computerized spermatogenesis staging (CSS) of mouse testis sections via quantitative histomorphological analysis. Med Image Anal. 2021;70:101835. https://doi.org/10.1016/j.media.2020.101835.
Kao C-Y, McMillan L. A novel deep learning architecture for testis histology image classification. arXiv preprint arXiv:1707.05809; 2017. https://doi.org/10.48550/arXiv.1707.05809.
Creasy DM, Panchal ST, Garg R, Samanta P. Deep learning-based spermatogenic staging assessment for hematoxylin and eosin-stained sections of rat testes. Toxicol Pathol. 2021;49(4):872–87. https://doi.org/10.1177/0192623320969678.
Liang S, Lu H, Zang M, Wang X, Jiao Y, Zhao T, Xu EY, Xu J. Deep SED-Net with interactive learning for multiple testicular cell types segmentation and cell composition analysis in mouse seminiferous tubules. Cytometry A. 2022. https://doi.org/10.1002/cyto.a.24556.
Ito Y, Unagami M, Yamabe F, Mitsui Y, Nakajima K, Nagao K, Kobayashi H. A method for utilizing automated machine learning for histopathological classification of testis based on Johnsen scores. Sci Rep. 2021;11(1):9962. https://doi.org/10.1038/s41598-021-89369-z.
Ghoshal B, Hikmet F, Pineau C, Tucker A, Lindskog C. DeepHistoClass: a novel strategy for confident classification of immunohistochemistry images using deep learning. Mol Cell Proteomics. 2021;20:100140. https://doi.org/10.1016/j.mcpro.2021.100140.
Riordon J, McCallum C, Sinton D. Deep learning for the classification of human sperm. Comput Biol Med. 2019;111:103342. https://doi.org/10.1016/j.compbiomed.2019.103342.
Movahed RA, Mohammadi E, Orooji M. Automatic segmentation of sperm’s parts in microscopic images of human semen smears using concatenated learning approaches. Comput Biol Med. 2019;109:242–53. https://doi.org/10.1016/j.compbiomed.2019.04.032.
Javadi S, Mirroshandel SA. A novel deep learning method for automatic assessment of human sperm images. Comput Biol Med. 2019;109:182–94. https://doi.org/10.1016/j.compbiomed.2019.04.030.
Bilgic E, Guzel E, Kose S, Aydin MC, Karaismailoglu E, Akar I, Usubutun A, Korkusuz P. Endocannabinoids modulate apoptosis in endometriosis and adenomyosis. Acta Histochem. 2017;119(5):523–32. https://doi.org/10.1016/j.acthis.2017.05.005.
Tzutalin, LabelImg. 2015. https://github.com/tzutalin/labelImg.
Roboflow. Roboflow. 2020; https://roboflow.com/about.
Ibtisham F, Wu J, Xiao M, An L, Banker Z, Nawab A, Zhao Y, Li G. Progress and future prospect of in vitro spermatogenesis. Oncotarget. 2017;8(39):66709–27. https://doi.org/10.18632/oncotarget.19640.
Sziva RE, Ács J, Tőkés AM, Korsós-Novák Á, Nádasy GL, Ács N, Horváth PG, Szabó A, Ke H, Horváth EM, Kopa Z, Várbíró S. Accurate quantitative histomorphometric-mathematical image analysis methodology of rodent testicular tissue and its possible future research perspectives in andrology and reproductive medicine. Life. 2022;12(2). https://doi.org/10.3390/life12020189.
Redmon J, Divvala S, Girshick R, Farhadi A. You only look once: unified, real-time object detection. In: Proceed IEEE Conference Comput Vision Pattern Recognition; 2016. https://doi.org/10.1109/CVPR.2016.91.
Fayomi AP, Orwig KE. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018;29:207–14. https://doi.org/10.1016/j.scr.2018.04.009.
Silber S. Histology of the testis and spermatogenesis. In: Silber S, editor. Fundamentals of Male Infertility. Cham: Springer International Publishing; 2018. p. 29–37. https://doi.org/10.1007/978-3-319-76523-5.
Du Y, Zhang R, Zargari A, Thai TC, Gunderson CC, Moxley KM, Liu H, Zheng B, Qiu Y. Classification of tumor epithelium and stroma by exploiting image features learned by deep convolutional neural networks. Ann Biomed Eng. 2018;46(12):1988–99. https://doi.org/10.1007/s10439-018-2095-6.
Wu M, Yan C, Liu H, Liu Q. Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks. Biosci Rep. 2018;38(3). https://doi.org/10.1042/BSR20180289.
Funding
This work was financially supported by Hacettepe University Scientific Research Project Coordination Unit; the (#TYL-2019-18375) TUBİTAK (The Scientific and Technological Research Council of Turkey) 1001 program supported Burak Kahveci as a scholar (#218S421).
Author information
Authors and Affiliations
Contributions
Burak Kahveci and Petek Korkusuz generated the hypothesis. Sections were taken by Selin Önen and Burak Kahveci. Selin Önen guided and they both performed the histochemical and immunohistochemical staining. Burak Kahveci created the datasets. Burak Kahveci and Fuat Akal created the DL models and carried out the experiments. Petek Korkusuz and Fuat Akal edited the manuscript. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Animal materials used in the experiments were approved by Hacettepe University Animal Experiments Local Ethics Committee (#2018, 52338575-96).
Consent to participate
Not applicable .
Consent for publication
Not applicable .
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kahveci, B., Önen, S., Akal, F. et al. Detection of spermatogonial stem/progenitor cells in prepubertal mouse testis with deep learning. J Assist Reprod Genet 40, 1187–1195 (2023). https://doi.org/10.1007/s10815-023-02784-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-023-02784-1