Abstract
Aim
To learn about the interaction between endometrial microbiota and host gene regulation in recurrent implantation failure.
Methods
The endometrial microbiota of 111 patients (RIF, 75; CON, 36) was analyzed by using 16 s rRNA sequencing technology. Transcriptome sequencing analysis of the endometrial of 60 patients was performed by using high-throughput sequencing.
Results
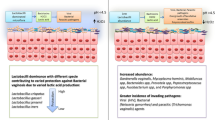
We found that the structure and composition of endometrium microbiota community of RIF patients were significantly different from those in control group. The abnormality of microbial structure and composition might interfere with the implantation of embryos by affecting the immune adaptation of the endometrium and the formation of endometrial blood vessels.
Conclusions
Our research described the host-microbe interaction in RIF. The structure and composition of endometrium microbiota community of RIF patients were significantly different from those in CON group. The abnormality of microbial structure and composition might interfere with the implantation of embryos by affecting the immune adaptation of the endometrium and the formation of endometrial blood vessels.





Similar content being viewed by others
Data availability
The 16 s rRNA gene sequencing for 75 endometrial microbiota samples have been deposited with the National Center for Biotechnology Information (NCBI) under reference number PRJNA732058. The transcriptome sequencing for 40 endometrium samples have been deposited with the NCBI under reference number PRJNA747622.
Abbreviations
- RIF:
-
Recurrent implantation failure
- ERA:
-
Endometrial receptivity array
- WOI:
-
Window of implantation
- FSH:
-
Follicle stimulation hormone
- PICRUSt2:
-
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- LEfSe:
-
Linear discriminant analysis effect size
References
Coughlan C, et al. Recurrent implantation failure: definition and management. Reprod Biomed Online. 2014;28(1):14–38.
Simon A, Laufer N. Repeated implantation failure: clinical approach. Fertil Steril. 2012;97(5):1039–43.
Thornhill AR, et al. ESHRE PGD Consortium “Best practice guidelines for clinical preimplantation genetic diagnosis (PGD) and preimplantation genetic screening (PGS)”. Hum Reprod. 2005;20(1):35–48.
Bashiri A, Halper KI, Orvieto R. Recurrent implantation failure-update overview on etiology, diagnosis, treatment and future directions. Reprod Biol Endocrinol. 2018;16(1):121.
Diaz-Gimeno P, et al. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil Steril. 2011;95(1):50–60, 60 e1–15.
Diaz-Gimeno P, et al. The accuracy and reproducibility of the endometrial receptivity array is superior to histology as a diagnostic method for endometrial receptivity. Fertil Steril. 2013;99(2):508–17.
Tan J, et al. The role of the endometrial receptivity array (ERA) in patients who have failed euploid embryo transfers. J Assist Reprod Genet. 2018;35(4):683–92.
Evans J, et al. Fertile ground: human endometrial programming and lessons in health and disease. Nat Rev Endocrinol. 2016;12(11):654–67.
Benner M, et al. How uterine microbiota might be responsible for a receptive, fertile endometrium. Hum Reprod Update. 2018;24(4):393–415.
Koedooder R, et al. Identification and evaluation of the microbiome in the female and male reproductive tracts. Hum Reprod Update. 2019;25(3):298–325.
Moreno I, et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol. 2016;215(6):684–703.
Molina NM, et al. New Opportunities for Endometrial Health by Modifying Uterine Microbial Composition: Present or Future? Biomolecules. 2020;10(4):593.
Chen C, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun. 2017;8(1):875.
O’Callaghan JL, et al. Re-assessing microbiomes in the low-biomass reproductive niche. BJOG. 2020;127(2):147–58.
Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6.
Rognes T, et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4: e2584.
Wang Q, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7.
Hill TC, et al. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol. 2003;43(1):1–11.
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. 2013;8(4): e61217.
Chong J, et al. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020;15(3):799–821.
Dhariwal A, et al. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45(W1):W180–8.
Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60.
Parks DH, et al. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–4.
Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8(9): e1002687.
Watts SC, et al. FastSpar: rapid and scalable correlation estimation for compositional data. Bioinformatics. 2019;35(6):1064–6.
Chen S, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60.
Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–5.
Pertea M, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–67.
Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7): e47.
Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Trygg J, Wold S. O2-PLS, a two-block (X–Y) latent variable regression (LVR) method with an integral OSC filter. J Chemom. 2003;17(1):53–64.
Bouhaddani SE, et al. Integrating omics datasets with the OmicsPLS package. BMC Bioinformatics. 2018;19(1):371.
Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Wei T, S.V. R package “corrplot”: visualization of a correlation matrix. 2021.
Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523.
Hashimoto T, Kyono K. Does dysbiotic endometrium affect blastocyst implantation in IVF patients? J Assist Reprod Genet. 2019;36(12):2471–9.
Teder H, et al. TAC-seq: targeted DNA and RNA sequencing for precise biomarker molecule counting. NPJ Genom Med. 2018;3:34.
Miao YR, et al. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv Sci (Weinh). 2020;7(7):1902880.
Molina NM, et al. Analysing endometrial microbiome: methodological considerations and recommendations for good practice. Hum Reprod. 2021;36(4):859–79.
Chaouat G. The Th1/Th2 paradigm: still important in pregnancy? Semin Immunopathol. 2007;29(2):95–113.
Candelli M, et al. Interaction between Lipopolysaccharide and Gut Microbiota in Inflammatory Bowel Diseases. Int J Mol Sci. 2021;22(12):6242.
Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–73.
Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10(4):311–23.
Koo HS, et al. CXCL12 enhances pregnancy outcome via improvement of endometrial receptivity in mice. Sci Rep. 2021;11(1):7397.
Koo HS, et al. Non-invasive intrauterine administration of botulinum toxin a enhances endometrial angiogenesis and improves the rates of embryo implantation. Reprod Sci. 2021;28(6):1671–87.
Corachán A, et al. Novel therapeutic targets to improve IVF outcomes in endometriosis patients: a review and future prospects. Hum Reprod Update. 2021;27(5):923–72.
Verstraelen H, et al. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1–2 region of the 16S rRNA gene. PeerJ. 2016;4: e1602.
Franasiak JM, et al. Endometrial microbiome at the time of embryo transfer: next-generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet. 2016;33(1):129–36.
Quince C, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6(9):639–41.
Huse SM, et al. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12(7):1889–98.
Quince C, et al. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38.
Mori H, et al. Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 2014;21(2):217–27.
Funding
National Natural Science Foundation of China [grant number 81871214] and National Key R&D Program of China [grant number 2017YFC1001603].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The Ethics Committee approved the study of Sixth Affiliated Hospital of Sun Yat-Sen University (L2021ZSLYEC-280).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Table S1
Clinical features of subject. (XLSX 17 kb)
Table S2
Differences in genus of RIF patients with different clinical characteristics. (XLSX 632 kb)

Figure S1
(A) The rarefaction curve of 16s rRNA data. The composition of Phylum (B) and Genus (C) of inter-individual in each group. (PNG 352 kb)

Figure S2
The co-occurrence network of RIF group (A) and CON group (B) based on SparCC. (PNG 473 kb)
Rights and permissions
About this article
Cite this article
Chen, P., Jia, L., Zhou, Y. et al. Interaction between endometrial microbiota and host gene regulation in recurrent implantation failure. J Assist Reprod Genet 39, 2169–2178 (2022). https://doi.org/10.1007/s10815-022-02573-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-022-02573-2