Abstract
Purpose
Approximately 1% of individuals who carry a balanced reciprocal translocation (BRT) are subfertile. Current karyotyping does not have the resolution to determine whether the breakpoints of the involved chromosomes perturb genes important for fertility. The aim of this study was to apply single-molecule optical mapping (SMOM) to patients presenting for IVF (in vitro fertilization) to ascertain whether the BRT disrupted any genes associated with normal fertility.
Methods
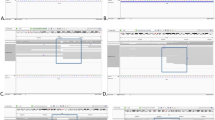
Nine subfertile patients with different BRTs were recruited for the study. Methyltransferase enzyme DLE1 was used to fluorescently label their genomic DNA samples at the recognition motif CTTAAG. The SMOM was performed on the Bionano platform, and long molecules aligned against the reference genome hg19 to identify the breakpoint regions. Mate-pair and PCR-Sanger sequencing were used to confirm the precise breakpoint sequences.
Results
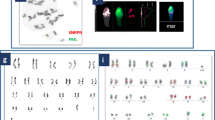
Both breakpoint regions in each of the nine BRTs were finely mapped to small regions of approximately 10 Kb, and their positions were consistent with original cytogenetic banding patterns determined by karyotyping. In three BRTs, breakpoints disrupted genes known to be associated with male infertility, namely NUP155 and FNDC3A [46,XY,t(5;13)(p15;q22)], DPY19L1 [46,XY,t(1;7)(p36.3;p15), and BAI3 [46,XY,t(3;6)(p21;q16)].
Conclusions
The SMOM has potential clinical application as a rapid tool to screen patients with BRTs for underlying genetic causes of infertility and other diseases.


Similar content being viewed by others
References
Ogilvie CM, Braude P, Scriven PN. Successful pregnancy outcomes after preimplantation genetic diagnosis (PGD) for carriers of chromosome translocations. Hum Fertil (Camb). 2001;4(3):168–71.
Wilch ES, Morton CC. Historical and clinical perspectives on chromosomal translocations. Adv Exp Med Biol. 2018;1044:1–14. https://doi.org/10.1007/978-981-13-0593-1_1.
Burrow AA, Williams LE, Pierce LC, Wang YH. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009;10:59. https://doi.org/10.1186/1471-2164-10-59.
Weckselblatt B, Rudd MK. Human structural variation: mechanisms of chromosome rearrangements. Trends Genet. 2015;31(10):587–99. https://doi.org/10.1016/j.tig.2015.05.010.
Nilsson D, Pettersson M, Gustavsson P, Forster A, Hofmeister W, Wincent J, et al. Whole-genome sequencing of cytogenetically balanced chromosome translocations identifies potentially pathological gene disruptions and highlights the importance of microhomology in the mechanism of formation. Hum Mutat. 2017;38(2):180–92. https://doi.org/10.1002/humu.23146.
Schluth-Bolard C, Labalme A, Cordier MP, Till M, Nadeau G, Tevissen H, et al. Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. J Med Genet. 2013;50(3):144–50. https://doi.org/10.1136/jmedgenet-2012-101351.
Kurahashi H, Kogo H, Tsutsumi M, Inagaki H, Ohye T. Failure of homologous synapsis and sex-specifc reproduction problems. Front Genet. 2012;3:112. https://doi.org/10.3389/fgene.2012.00112.
Fischer J, Colls P, Escudero T, Munne S. Preimplantation genetic diagnosis (PGD) improves pregnancy outcome for translocation carriers with a history of recurrent losses. Fertil Steril. 2010;94(1):283–9. https://doi.org/10.1016/j.fertnstert.2009.02.060.
Zhang W, Liu Y, Wang L, Wang H, Ma M, Xu M, et al. Clinical application of next-generation sequencing in preimplantation genetic diagnosis cycles for Robertsonian and reciprocal translocations. J Assist Reprod Genet. 2016;33(7):899–906. https://doi.org/10.1007/s10815-016-0724-2.
Wang L, Shen J, Cram DS, Ma M, Wang H, Zhang W, et al. Preferential selection and transfer of euploid noncarrier embryos in preimplantation genetic diagnosis cycles for reciprocal translocations. Fertil Steril. 2017;108(4):620–7 e4. https://doi.org/10.1016/j.fertnstert.2017.07.010.
Treff NR, Thompson K, Rafizadeh M, Chow M, Morrison L, Tao X, et al. SNP array-based analyses of unbalanced embryos as a reference to distinguish between balanced translocation carrier and normal blastocysts. J Assist Reprod Genet. 2016;33(8):1115–9. https://doi.org/10.1007/s10815-016-0734-0.
Trask BJ. Human cytogenetics: 46 chromosomes, 46 years and counting. Nat Rev Genet. 2002;3(10):769–78. https://doi.org/10.1038/nrg905.
Wapner RJ, Martin CL, Levy B, Ballif BC, Eng CM, Zachary JM, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367(23):2175–84. https://doi.org/10.1056/NEJMoa1203382.
Carpenter NJ. Molecular cytogenetics. Semin Pediatr Neurol. 2001;8(3):135–46.
Aristidou C, Koufaris C, Theodosiou A, Bak M, Mehrjouy MM, Behjati F, et al. Accurate breakpoint mapping in apparently balanced translocation families with discordant phenotypes using whole genome mate-pair sequencing. PLoS One. 2017;12(1):e0169935. https://doi.org/10.1371/journal.pone.0169935.
Liang D, Wang Y, Ji X, Hu H, Zhang J, Meng L, et al. Clinical application of whole-genome low-coverage next-generation sequencing to detect and characterize balanced chromosomal translocations. Clin Genet. 2017;91(4):605–10. https://doi.org/10.1111/cge.12844.
Chan EKF, Cameron DL, Petersen DC, Lyons RJ, Baldi BF, Papenfuss AT, et al. Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 2018;28(5):726–38. https://doi.org/10.1101/gr.227975.117.
Levy-Sakin M, Pastor S, Mostovoy Y, Li L, Leung AKY, McCaffrey J, et al. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun. 2019;10(1):1025. https://doi.org/10.1038/s41467-019-08992-7.
Zhang Q, Xu X, Ding L, Li H, Xu C, Gong Y, et al. Clinical application of single-molecule optical mapping to a multigeneration FSHD1 pedigree. Mol Genet Genomic Med. 2019;7(3):e565. https://doi.org/10.1002/mgg3.565.
Obholz KL, Akopyan A, Waymire KG, MacGregor GR. FNDC3A is required for adhesion between spermatids and Sertoli cells. Dev Biol. 2006;298(2):498–513. https://doi.org/10.1016/j.ydbio.2006.06.054.
Gigliotti S, Callaini G, Andone S, Riparbelli MG, Pernas-Alonso R, Hoffmann G, et al. Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the nup155 vertebrate nucleoporin gene. J Cell Biol. 1998;142(5):1195–207. https://doi.org/10.1083/jcb.142.5.1195.
Ray PF, Coutton C, Arnoult C. Sun proteins and Dpy19l2 forming LINC-like links are critical for spermiogenesis. Biol Open. 2016;5(5):535–6. https://doi.org/10.1242/bio.016626.
Koscinski I, Elinati E, Fossard C, Redin C, Muller J, Velez de la Calle J, et al. DPY19L2 deletion as a major cause of globozoospermia. Am J Hum Genet. 2011;88(3):344–50. https://doi.org/10.1016/j.ajhg.2011.01.018.
Bansal SK, Gupta N, Sankhwar SN, Rajender S. Differential genes expression between fertile and infertile spermatozoa revealed by transcriptome analysis. PLoS One. 2015;10(5):e0127007. https://doi.org/10.1371/journal.pone.0127007.
Cram DS, O’Bryan MK, de Kretser DM. Male infertility genetics--the future. J Androl. 2001;22(5):738–46.
Cram D, Lynch M, O’Bryan MK, Salvado C, McLachlan RI, de Kretser DM. Genetic screening of infertile men. Reprod Fertil Dev. 2004;16(5):573–80. https://doi.org/10.10371/RD03097.
Harton GL, Tempest HG. Chromosomal disorders and male infertility. Asian J Androl. 2012;14(1):32–9. https://doi.org/10.1038/aja.2011.66.
Chantot-Bastaraud S, Ravel C, Siffroi JP. Underlying karyotype abnormalities in IVF/ICSI patients. Reprod BioMed Online. 2008;16(4):514–22.
Stern C, Pertile M, Norris H, Hale L, Baker HW. Chromosome translocations in couples with in-vitro fertilization implantation failure. Hum Reprod. 1999;14(8):2097–101. https://doi.org/10.1093/humrep/14.8.2097.
Schreurs A, Legius E, Meuleman C, Fryns JP, D’Hooghe TM. Increased frequency of chromosomal abnormalities in female partners of couples undergoing in vitro fertilization or intracytoplasmic sperm injection. Fertil Steril. 2000;74(1):94–6. https://doi.org/10.1016/s0015-0282(00)00558-6.
Scriven PN, Flinter FA, Khalaf Y, Lashwood A, Ogilvie CM. Benefits and drawbacks of preimplantation genetic diagnosis (PGD) for reciprocal translocations: lessons from a prospective cohort study. Eur J Hum Genet. 2013;21(10):1035–41. https://doi.org/10.1038/ejhg.2013.9.
Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet. 1991;49(5):995–1013.
Vasilevska M, Ivanovska E, Kubelka Sabit K, Sukarova-Angelovska E, Dimeska G. The incidence and type of chromosomal translocations from prenatal diagnosis of 3800 patients in the republic of Macedonia. Balkan J Med Genet. 2013;16(2):23–8. https://doi.org/10.2478/bjmg-2013-0027.
Acknowledgments
The authors thank the patients who participated in this research study.
Funding
This work was supported by National Key Research and Development Program of China (2018YFC1003100).
Author information
Authors and Affiliations
Contributions
H.W. and B.X.: study design and management and supervision of the study. A.M. and D.S.C.: study design, statistical analysis, and manuscript preparation. Z.J., A.M., H.Z., and T.Y.: experimental procedures and statistical analysis. X.Z. and T.M.: data analysis. S.W., L.W., and S.L.: supervision. M.X.: data analysis and manuscript preparation. Y.Y.: supervision and manuscript review.
Corresponding authors
Ethics declarations
Conflict of interest
A.M., H.Z., X.Z., T.Y, T.M., M.X., and D.S.C. are employees of Berry Genomics Corporation. None of the authors holds any stocks or bonds in the company. The other authors declare that they have no conflict of interest.
Ethical approval
Ethical approval for performing this study was obtained from the Ethics Committee of the Chinese PLA General Hospital (Approval number S2013-092-02).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 13.6 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Jia, Z., Mao, A. et al. Analysis of balanced reciprocal translocations in patients with subfertility using single-molecule optical mapping. J Assist Reprod Genet 37, 509–516 (2020). https://doi.org/10.1007/s10815-020-01702-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-020-01702-z