Abstract
Purpose
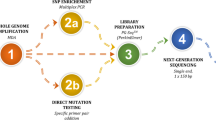
Development of PGD assays for molecular disorders is based on analysis of a familial mutation together with linked polymorphic STR markers; a process which is lengthy and requires the identification of multiple informative markers prior to PGD analysis. On the other hand, whole genome amplification (WGA), in conjunction with microarray platforms, allows the use of a universal assay for the analysis of a very large number of SNP markers at once. The aim of this study was to test high throughput pre-PGD familial haplotyping for in-case blastomere analysis in order to eliminate time-consuming pre-case preparations for each family.
Methods
A PGD cycle was performed for a couple with paternal Charcot Marie Tooth 1A (CMT1A) using a classic multiplex nested PCR approach. Mutant embryos from the case were blindly reanalyzed, as single or multi-cell biopsies, using a multiple displacement amplification-based WGA protocol and microarray SNP analysis. In parallel, relevant genomic DNA samples from the family were also analyzed by SNP microarray.
Results
After applying a ‘unique informative allele’ selection algorithm to the data, this array-based assay reconfirmed the initial diagnosis in all samples.
Conclusions
We describe a PGD method that is both accurate and feasible during the time-frame required for embryo transfer. This strategy greatly reduces the time for pre-case haplotype preparation.





Similar content being viewed by others
References
Altarescu G, Brooks B, Kaplan Y, Eldar-Geva T, Margalioth EJ, Levy-Lahad E, et al. Single-sperm analysis for haplotype construction of de-novo paternal mutations: application to PGD for neurofibromatosis type 1. Hum Reprod. 2006;21(8):2047–51. doi:10.1093/humrep/del064.
Altarescu G, Eldar Geva T, Brooks B, Margalioth E, Levy-Lahad E, Renbaum P. PGD on a recombinant allele: crossover between the TSC2 gene and ‘linked’ markers impairs accurate diagnosis. Prenat Diagn. 2008;28(10):929–33. doi:10.1002/pd.2070.
Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009;5(9):e1000648. doi:10.1371/journal.pgen.1000648.
Fragouli E, Wells D. Aneuploidy screening for embryo selection. Sem Reprod Med. 2012;30(4):289–301. doi:10.1055/s-0032-1313908.
Handyside AH, Harton GL, Mariani B, Thornhill AR, Affara N, Shaw MA, et al. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. 2010;47(10):651–8. doi:10.1136/jmg.2009.069971.
Handyside AH, Kontogianni EH, Hardy K, Winston RM. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature. 1990;344(6268):768–70. doi:10.1038/344768a0.
Iwamoto K, Bundo M, Ueda J, Nakano Y, Ukai W, Hashimoto E, et al. Detection of chromosomal structural alterations in single cells by SNP arrays: a systematic survey of amplification bias and optimized workflow. PLoS One. 2007;2(12):e1306. doi:10.1371/journal.pone.0001306.
Jin R, Tong X, Wu L, Luo L, Luan H, Zhou G, et al. Extended culture of vitrified-warmed embryos in day-3 embryo transfer cycles: a randomized controlled pilot study. Reprod Biomed Online. 2013;26(4):384–92. doi:10.1016/j.rbmo.2012.12.003.
Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod. 2010;25(4):1066–75. doi:10.1093/humrep/dep452.
Kakourou G, Dhanjal S, Daphnis D, Doshi A, Nuttall S, Gotts S, et al. Preimplantation genetic diagnosis for myotonic dystrophy type 1: detection of crossover between the gene and the linked marker APOC2. Prenat Diagn. 2007;27(2):111–6. doi:10.1002/pd.1611.
Konings P, Vanneste E, Jackmaert S, Ampe M, Verbeke G, Moreau Y, et al. Microarray analysis of copy number variation in single cells. Nat Protoc. 2012;7(2):281–310. doi:10.1038/nprot.2011.426.
Renwick PJ, Trussler J, Ostad-Saffari E, Fassihi H, Black C, Braude P, et al. Proof of principle and first cases using preimplantation genetic haplotyping–a paradigm shift for embryo diagnosis. Reprod Biomed Online. 2006;13(1):110–9.
Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I, et al. Optimization and evaluation of single-cell whole-genome multiple displacement amplification. Hum Mutat. 2006;27(5):496–503. doi:10.1002/humu.20324.
Treff NR, Su J, Tao X, Northrop LE, Scott Jr RT. Single-cell whole-genome amplification technique impacts the accuracy of SNP microarray-based genotyping and copy number analyses. Mol Hum Reprod. 2011;17(6):335–43. doi:10.1093/molehr/gaq103.
Vanneste E, Bittman L, Van der Aa N, Voet T, Vermeesch JR. New array approaches to explore single cells genomes. Front Genet. 2012;3:44. doi:10.3389/fgene.2012.00044.
Verlinsky O, Kuliev A. Atlas of preimplantation genetic diagnosis. 2nd ed. London: Taylor & Francis; 2004.
Verlinsky Y, Cohen J, Munne S, Gianaroli L, Simpson JL, Ferraretti AP, et al. Over a decade of experience with preimplantation genetic diagnosis. Fertil Steril. 2004;82(2):302–3. doi:10.1016/j.fertnstert.2004.02.108.
Voet T, Vanneste E, Van der Aa N, Melotte C, Jackmaert S, Vandendael T, et al. Breakage-fusion-bridge cycles leading to inv dup del occur in human cleavage stage embryos. Hum Mutat. 2011;32(7):783–93. doi:10.1002/humu.21502.
Wilton L, Thornhill A, Traeger-Synodinos J, Sermon KD, Harper JC. The causes of misdiagnosis and adverse outcomes in PGD. Hum Reprod. 2009;24(5):1221–8. doi:10.1093/humrep/den488.
Zhu D, Zhang J, Cao S, Zhang J, Heng BC, Huang M, et al. Vitrified-warmed blastocyst transfer cycles yield higher pregnancy and implantation rates compared with fresh blastocyst transfer cycles-time for a new embryo transfer strategy? Fertil Steril. 2011;95(5):1691–5. doi:10.1016/j.fertnstert.2011.01.022.
Acknowledgments
We thank Rabbi David and Mrs. Anita Fuld for their generous and ongoing support.
Conflicts of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Capsule The classic STR-based method for PGD of monogenic disorders requires time consuming pre-case preparations. In this proof of principle investigation, we describe a SNP microarray-based methodology that accomplishes pre-PGD case preparations in a fraction of the time without compromising on diagnostic accuracy.
G. Altarescu and D. A. Zeevi are equal contributing authors
Rights and permissions
About this article
Cite this article
Altarescu, G., Zeevi, D.A., Zeligson, S. et al. Familial haplotyping and embryo analysis for Preimplantation Genetic Diagnosis (PGD) using DNA microarrays: a proof of principle study. J Assist Reprod Genet 30, 1595–1603 (2013). https://doi.org/10.1007/s10815-013-0044-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-013-0044-8