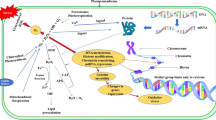
Abstract
Kappaphycus alvarezii is an important kappa-carrageenan producer. Fluctuating environmental conditions, especially seawater salinity, significantly affect the cultivation of K. alvarezii. To assess the variable salinity-induced responses, biochemical responses and gene expressions via high throughput RNA sequencing were studied in K. alvarezii under hypo- [3.25 practical salinity units (psu)] and hyper- (65 psu) salinity conditions. Under variable salinity, the influx of Na+ and efflux of K+ ions resulted in a lower K+/Na+ ratio. Both salinity levels had differential effect on accumulation of different solutes and photosynthetic pigments. Superoxide dismutase, ascorbate peroxidase and glutathione peroxidase exhibited higher activity under stress. The RNA sequencing revealed the differential gene expression in K. alvarezii at variable salinity. In this study, 52,875 CDS were predicted, and most of these showed a match with Chondrus crispus. At hypo-salinity, 246 and 171 genes exhibited up- and down-regulation, respectively. At hyper-salinity, 457 and 1,008 exhibited up- and down-regulation, respectively. Transcriptome data revealed differential expression of genes encoding components of photosynthetic, signal transduction, fatty acid and amino acid metabolism, energy metabolism and redox homeostasis machineries under stress. Differential expression of ubiquitin-mediated proteolytic genes and heat shock proteins indicated the important role of these genes under studied strengths of salinity. The differentially expressed genes showed a correlation with biochemical responses and ion accumulation. The study identified numerous key genes regulating different metabolic pathways and antioxidant machinery. The results would facilitate insights into the molecular mechanism underlying the salinity stress adaptation in K. alvarezii and studies on the gene discoveries for subsequent strain improvement.









Similar content being viewed by others
Data Availability
Transcriptome data are available on NCBI domain with accession number PRJNA661401. This information may please be provided as data availability statement.
References
Agrawal S, Siddiqui SA, Chaudhary DR, Rathore MS (2022) Bio-prospection and compositional multivariate analysis revealed the industrial potential of selected seaweeds along Gujarat seacoast. Bioresour Technol Rep 19:101130
Beyer WF, Fridovich I (1987) Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Ana Biochem 161:559–566
Bischof K, Rautenberger R (2012) Seaweed responses to environmental stress: reactive oxygen and antioxidative strategies. In: Bishof K, Wiencke C (eds) Seaweed Biology. Springer, Heidelberg, pp 109–132
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Collén J, Guisle-Marsollier I, Léger JJ, Boyen C (2007) Response of the transcriptome of the intertidal red seaweed Chondrus crispus to controlled and natural stresses. New Phytol 176:45–55
Collén J, Pedersén M, Bornman CH (1994) A stress-induced oxidative burst in Eucheuma platycladum (Rhodophyta). Physiol Plant 92:417–422
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Contreras-Porcia L, López-Cristoffanini C, Lovazzano C, Flores-Molina MR, Thomas D, Núñez A, Fierro C, Guajardo E, Correa JA, Kube M, Reinhardt R (2013) Differential gene expression in Pyropia columbina (Bangiales, Rhodophyta) under natural hydration and desiccation conditions. Lat Am J Aquat Res 41:933–958
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11:163
Das P, Mondal D, Maiti S (2017) Thermochemical conversion pathways of Kappaphycus alvarezii granules through study of kinetic models. Bioresour Technol 234:233–242
De Marsac NT, Houmard J (1988) Complementary chromatic adaptation: Physiological conditions and action spectra. Meth Enzymol 167:318–328
Dittami SM, Scornet D, Petit JL, Ségurens B, da Silva C, Corre E, Dondrup M, Glatting KH, König R, Sterck L, Rouzé P, van de Peer Y, Cock JM, Boyen C, Tonon T (2009) Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress. Genome Biol 10:R66
Dring MJ (2006) Stress resistance and disease resistance in seaweeds: the role of reactive oxygen metabolism. Adv Bot Res 43:175–207
Fattorusso E, Piatelli M (1980) Amino acids from marine algae. In: Scheuer PJ (ed) Marine natural products: Chemical and biological perspectives, Vol III. Academic Press, Toronto pp 95–140
Gao S, Niu J, Chen W, Wang G, Xie X, Pan G, Gu W, Zhu D (2013) The physiological links of the increased photosystem II activity in moderately desiccated Porphyra haitanensis (Bangiales, Rhodophyta) to the cyclic electron flow during desiccation and re-hydration. Photosynth Res 116:45–54
Gigon A, Matos AR, Laffray D, Zuily-Fodil Y, Pham-Thi AT (2004) Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia). Ann Bot 94:345–351
Góes HG, Reis RP (2011) An initial comparison of tubular netting versus tie-tie methods of cultivation for Kappaphycus alvarezii (Rhodophyta, Solieriaceae) on the south coast of Rio de Janeiro State, Brazil. J Appl Phycol 23:607–613
Goyal A (2007) Osmoregulation in Dunaliella, Part II: Photosynthesis and starch contribute carbon for glycerol synthesis during a salt stress in Dunaliella tertiolecta. Plant Physiol Biochem 45:705–710
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, ouger MB, Eccles D, Li B, Lieber M, MacManes MD (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Prot 8:1494
Jacob PT, Siddiqui SA, Rathore MS (2020) Seed germination, seedling growth and seedling development associated physio-chemical changes in Salicornia brachiata (Roxb.) under salinity and osmotic stress. Aquat Bot 166:103272
Jebara S, Jebara M, Limam F, Aouani ME (2005) Changes in ascorbate peroxidase, catalase, guaiacol peroxidase and superoxide dismutase activities in common bean (Phaseolus vulgaris) nodules under salt stress. J Plant Physiol 162:929–936
Ji Z, Zou D, Gong J, Liu C, Ye C, Chen Y (2019) The different responses of growth and photosynthesis to NH4+ enrichments between Gracilariopsis lemaneiformis and its epiphytic alga Ulva lactuca grown at elevated atmospheric CO2. Mar Pollut Bull 144:173–180
Jia Z, Niu J, Huan L, Wu X, Wang G, Hou Z (2013) Cyclophilin participates in responding to stress situations in Porphyra haitanensis (Bangiales, Rhodophyta). J Phycol 49:194–201
Junglee S, Urban L, Sallanon H, Lopez-Lauri F (2014) Optimized assay for hydrogen peroxide determination in plant tissue using potassium iodide. Am J Anal Chem 5:730–736
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280
Karsten U (2007) Salinity tolerance of Arctic kelps from Spitsbergen. Phycol Res 55:257–262
Kerjean V, Morel B, Stiger V, Bessieres MA, Simon-Colin C, Magné C, Deslandes E (2007) Optimization of floridoside production in the red alga Mastocarpus stellatus: Pre treatment, extraction and seasonal variations. Bot Mar 50:59–64
Khatri K, Rathore MS (2022) Salt and osmotic stress-induced changes in physio-chemical responses, PSII photochemistry and chlorophyll a fluorescence in peanut. Plant Stress 3:100063
Kim JG, Back K, Lee HY, Lee HJ, Phung TH, Grimm B, Jung S (2014) Increased expression of Fe-chelatase leads to increased metabolic flux into heme and confers protection against photodynamically induced oxidative stress. Plant Mol Biol 86:271–287
Kumar M, Kumari P, Gupta V, Reddy CR, Jha B (2010) Biochemical responses of red alga Gracilaria corticata (Gracilariales, Rhodophyta) to salinity induced oxidative stress. J Exp Mar Biol Ecol 391:27–34
Kumari J, Haque M, Jha RK, Rathore MS (2022) The red seaweed Kappaphycus alvarezii antiporter gene (KaNa+/H+) confers abiotic stress tolerance in transgenic tobacco. Mol Biol Rep 49:3729–3743
Lalegerie F, Gager L, Stiger-Pouvreau V, Connan S (2020) The stressful life of red and brown seaweeds on the temperate intertidal zone: Effect of abiotic and biotic parameters on the physiology of macroalgae and content variability of particular metabolites. Adv Bot Res 95:247–287
Lee WK, Namasivayam P, Ong Abdullah J, Ho CL (2017) Transcriptome profiling of sulfate deprivation responses in two agarophytes Gracilaria changii and Gracilaria salicornia (Rhodophyta). Sci Rep 7: srep46563.
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li Q, Zhang L, Pang T, Liu J (2019) Comparative transcriptome profiling of Kappaphycus alvarezii (Rhodophyta, Gigartinales) in response to two extreme temperature treatments: an RNA-seq-based resource for photosynthesis research. Eur J Phycol 54:162–174
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Liu CL, Wang XL, Huang XH, Liu J (2011) Identification of hypo-osmotically induced genes in Kappachycus alvarezii (Solieriaceae, Rhodophyta) through expressed sequence tag analysis. Bot Mar 54:557–562
Lu IF, Sung MS, Lee TM (2006) Salinity stress and hydrogen peroxide regulation of antioxidant defense system in Ulva fasciata. Mar Biol 150:1–15
Lu X, Huan L, Gao S, He L, Wang G (2016) NADPH from the oxidative pentose phosphate pathway drives the operation of cyclic electron flow around photosystem I in high-intertidal macroalgae under severe salt stress. Physiol Plant 156:397–406
Luo MB, Liu F (2011) Salinity-induced oxidative stress and regulation of antioxidant defense system in the marine macroalga Ulva prolifera. J Exp Mar Biol Ecol 409:223–228
Macler BA (1988) Salinity effects on photosynthesis, carbon allocation and nitrogen assimilation in the red alga Gelidium coulteri. Plant Physiol 88:690–694
Mandal SK, Ajay G, Monisha N, Malarvizhi J, Temkar G, Mantri VA (2015) Differential response of varying temperature and salinity regimes on nutrient uptake of drifting fragments of Kappaphycus alvarezii: implication on survival and growth. J Appl Phycol 27:1571–1581
Miyagawa Y, Tamoi M, Shigeoka S (2000) Evaluation of the defense system in chloroplasts to photooxidative stress caused by paraquat using transgenic tobacco plants expressing catalase from Escherichia coli. Plant Cell Physiol 41:311–320
Mohanty B, Kitazumi A, Cheung CM, Lakshmanan M, Benildo G, Jang IC, Lee DY (2016) Identification of candidate network hubs involved in metabolic adjustments of rice under drought stress by integrating transcriptome data and genome-scale metabolic network. Plant Sci 242:224–239
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185
Muzi C, Camoni L, Visconti S, Aducci P (2016) Cold stress affects H+-ATPase and phospholipase D activity in Arabidopsis. Plant Physiol Biochem 108:328–336
Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22:867–880
Narusak Y, Narusaka M, Seki M, Umezawa T, Ishida J, Nakajima M, Enju A, Shinozaki K (2004) Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol Biol 55:327–342
Obata T, Fernie AR (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69:3225–3243
Pan L, Zhang X, Wang J, Ma X, Zhou M, Huang L, Nie G, Wang P, Yang Z, Li J (2016) Transcriptional profiles of drought-related genes in modulating metabolic processes and antioxidant defenses in Lolium multiflorum. Front Plant Sci 7:519
Pereira DT, Simioni C, Filipin EP, Bouvie F, Ramlov F, Maraschin M, Bouzon ZL, Schmidt ÉC (2017) Effects of salinity on the physiology of the red macroalga, Acanthophora spicifera (Rhodophyta, Ceramiales). Acta Bot Bras 31:555–565
Pliego-Cortés H, Caamal-Fuentes E, Montero-Muñoz J, Freile-Pelegrín Y, Robledo D (2017) Growth, biochemical and antioxidant content of Rhodymenia pseudopalmata (Rhodymeniales, Rhodophyta) cultivated under salinity and irradiance treatments. J Appl Phycol 29:2595–2603
Reed RH, Collins JC, Russell G (1981) The Effects of Salinity upon Ion Content and Ion Transport of the Marine Red Alga Porphyra purpurea (Roth) C. Ag J Exp Bot 32:347–367
Samanta P, Shin S, Jang S, Kim JK (2019) Comparative assessment of salinity tolerance based on physiological and biochemical performances in Ulva australis and Pyropia yezoensis. Algal Res 42:101590
Sampath-Wiley P, Neefus CD (2007) An improved method for estimating R-phycoerythrin and R-phycocyanin contents from crude aqueous extracts of Porphyra (Bangiales, Rhodophyta). J Appl Phycol 19:123–129
Scherer GE (2010) Phospholipase A in plant signal transduction. In: Munnik T (ed) Lipid Signaling in Plants. Springer, Heidelberg pp 3–22
Schopfer P, Plachy C, Frahry G (2001) Release of reactive oxygen intermediates (superoxide radicals, hydrogen peroxide, and hydroxyl radicals) and peroxidase in germinating radish seeds controlled by light, gibberellin, and abscisic acid. Plant Physiol 125:1591–1602
Siddiqui SA, Agrawal S, Brahmbhatt H, Rathore MS (2022) Metabolite expression changes in Kappaphycus alvarezii (a red alga) under hypo-and hyper-saline conditions. Algal Res 63:102650
Smirnoff N, Arnaud D (2018) Hydrogen peroxide metabolism and functions in plants. New Phytol 221:1197–1214
Smith IK, Vierheller TL, Thorne CA (1988) Assay of glutathione reductase in crude tissue homogenates using 5,5′-dithiobis(2-nitrobenzoic acid). Anal Biochem 175:408–413
Song L, Wu S, Sun J, Wang L, Liu T, Chi S, Liu C, Li X, Yin J, Wang X, Yu J (2014) De novo sequencing and comparative analysis of three red algal species of family Solieriaceae to discover putative genes associated with carrageenan biosynthesis. Acta Oceanol Sin 33:45–53
Sun P, Mao Y, Li G, Cao M, Kong F, Wang L, Bi G (2015) Comparative transcriptome profiling of Pyropia yezoensis (Ueda) M.S. Hwang and H.G. Choi in response to temperature stresses. BMC Genom 16:463
Sung MS, Hsu YT, Hsu YT, Wu TM, Lee TM (2009) Hypersalinity and hydrogen peroxide upregulation of gene expression of antioxidant enzymes in Ulva fasciata against oxidative stress. Mar Biotechnol 11:199–209
Surget G, Le Lann K, Delebecq G, Kervarec N, Donval A, Poullaouec MA, Bihannic I, Poupart N, Stiger-Pouvreau V (2017) Seasonal phenology and metabolomics of the introduced red macroalga Gracilaria vermiculophylla, monitored in the Bay of Brest (France). J Appl Phycol 29:2651–2666
Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15:462–470
Tammam AA, Fakhry EM, El-Sheekh M (2011) Effect of salt stress on antioxidant system and the metabolism of the reactive oxygen species in Dunaliella salina and Dunaliella tertiolecta. Afr J Biotechnol 10:3795–3808
Teo SS, Ho CL, Teoh S, Rahim RA, Phang SM (2009) Transcriptomic analysis of Gracilaria changii (Rhodophyta) in response to hyper- and hypoosmotic stresses. J Phycol 45:1093–1099
Thalmann M, Santelia D (2017) Starch as a determinant of plant fitness under abiotic stress. New Phytol 214:943–951
Thien VY, Rodrigues KF, Voo CLY, Wong CMVL, Yong WTL (2021) Comparative transcriptome profiling of Kappaphycus alvarezii (Rhodophyta, Solieriaceae) in response to light of different wavelengths and carbon dioxide enrichment. Plants 10:1236
Touchette BW (2007) Seagrass-salinity interactions: Physiological mechanisms used by submersed marine angiosperms for a life at sea. J Exp Mar Biol Ecol 350:194–215
Vairappan C, Chung C, Hurtado A, Soya F, Lhonneur G, Critchley A (2008) Distribution and symptoms of epiphyte infection in major carrageenophyte-producing farms. J Appl Phycol 20:477–483
van Ginneken V (2018) Some mechanism seaweeds employ to cope with salinity stress in the harsh euhaline oceanic environment. Am J Plant Sci 9:1191–1211
Vorphal MA, Gallardo-Escárate C, Valenzuela-Muñoz V, Dagnino-Leone J, Vásquez JA, Martínez-Oyanedel J, Bunster M (2017) De novo transcriptome analysis of the red seaweed Gracilaria chilensis and identification of linkers associated with phycobilisomes. Mar Genom 31:17–19
Wang W, Lin Y, Teng F, Ji D, Xu Y, Chen C, Xie C (2018) Comparative transcriptome analysis between heat-tolerant and sensitive Pyropia haitanensis strains in response to high temperature stress. Algal Res 29:104–112
Wang W, Shen Z, Sun X, Liu F, Liang Z, Wang F, Zhu J (2019a) De novo transcriptomics analysis revealed a global reprogramming towards dehydration and hyposalinity in Bangia fuscopurpurea gametophytes (Rhodophyta). J Appl Phycol 31:637–651
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252
Wang W, Xing L, Xu K, Ji D, Xu Y, Chen C, Xie C (2020) Salt stress-induced H2O2 and Ca2+ mediate K+/Na+ homeostasis in Pyropia haitanensis. J Appl Phycol 32:4199–4210
Wang WJ, Li XL, Zhu JY, Liang ZR, Liu FL, Sun XT, Wang FJ, Shen ZG (2019b) Antioxidant response to salinity stress in freshwater and marine Bangia (Bangiales, Rhodophyta). Aquat Bot 154:35–41
Wu Y, Jin X, Liao W, Hu L, Dawuda MM, Zhao X, Tang Z, Gong T, Yu J (2018) 5-Aminolevulinic acid (ALA) alleviated salinity stress in cucumber seedlings by enhancing chlorophyll synthesis pathway. Front Plant Sci 9:635
Xu K, Xu Y, Ji D, Xie J, Chen C, Xie C (2016) Proteomic analysis of the economic seaweed Pyropia haitanensis in response to desiccation. Algal Res 19:198–206
Yadav SK, Khatri K, Rathore MS, Jha B (2018) Introgression of UfCyt c6, a thylakoid lumen protein from a green seaweed Ulva fasciata Delile enhanced photosynthesis and growth in tobacco. Mole Biol Rep 45:1745–1758
Yadav SK, Khatri K, Rathore MS, Jha B (2020) Ectopic Expression of a Transmembrane protein KaCyt b6 from a red seaweed Kappaphycus alvarezii in transgenic tobacco augmented the photosynthesis and growth. DNA Cell Biol. https://doi.org/10.1089/dna.2020.5479
Yamasaki T, Yamakawa T, Yamane Y, Koike H, Satoh K. Katoh S (2002) Temperature acclimation of photosynthesis and related changes in photosystem II electron transport in winter wheat. Plant Physiol 128:1087–1097
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
Yee TV, Rodrigues KF, Lym WYT, Ling CMWV (2017) Comparison of gene expression patterns of Kappaphycus alvarezii (Rhodophyta, Solieriaceae) under different light wavelengths and CO2 enrichment. bioRxiv 188250
Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W (2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 188:762–773
Yunque DAT, Tibubos KR, Hurtado AQ, Critchley AT (2011) Optimization of culture conditions for tissue culture production of young plantlets of carrageenophyte Kappaphycus. J Appl Phycol 23:433–438
Zhang H, Li W, Li J, Fu W, Yao J, Duan D (2012) Characterization and expression analysis of hsp70 gene from Ulva prolifera J. Agardh (Chlorophycophyta, Chlorophyceae). Mar Genom 5:53–58
Zhang Z, Pang T, Li Q, Zhang L, Li L, Liu J (2015) Transcriptome sequencing and characterization for Kappaphycus alvarezii. Eur J Phycol 50:400–407
Acknowledgment
CSIR-CSMCRI PRIS: 129/2022
Authors are thankful to CSIR (Government of India), New Delhi for financial support under different plan projects. The authors are thankful to analytical facility of CSMCRI institute. KK thanks CSIR (Government of India), New Delhi for Junior/Senior Research Fellowship (JRF/SRF) and Academy of Scientific and Innovative Research (AcSIR), Ghaziabad for registration in Ph.D. program.
Author information
Authors and Affiliations
Contributions
MSR and SS- Conceptualization, fund raising, experimental design, editing and finalization of manuscript; KK- designing and execution of experiments, data analyses, drafting and editing of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Competing interest
Authors declare no potential conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10811_2023_3006_MOESM1_ESM.pptx
Fig. S1 Ion (a) and chlorophyll and carotenoids (b) contents in K. alvarezii under hypo- and hyper-saline conditions (PPTX 52.5 KB)
10811_2023_3006_MOESM4_ESM.pptx
Fig. S4 GO term distribution for biological functions (a), cellular components (b) and molecular functions (c) in pooled CDS of K. alvarezii (PPTX 58.6 KB)
10811_2023_3006_MOESM8_ESM.pdf
Table S2 Correlation matrix of different physio-biochemical activities and contents observed in Kappaphycus alvarezii under hypo- and hyper-saline conditions (PDF 138 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khatri, K., Sharma, S. & Rathore, M.S. De novo transcriptome assembly and variable salinity induced differential biochemical and transcript responses in Kappaphycus alvarezii, a red carrageenophyte. J Appl Phycol 35, 1847–1864 (2023). https://doi.org/10.1007/s10811-023-03006-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-023-03006-y