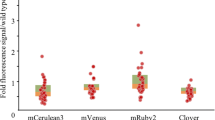
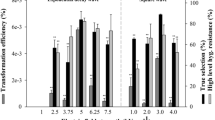
Abstract
Chlorella is a green unicellular alga that has a wide range of future biotechnical applications such as production of pharmaceuticals and biodiesel. Efficient genetic transformation of Chlorella vulgaris has been difficult due to technical limitations. In this study, an efficient and reliable transformation system of electroporation was established using two different reporter genes. First, C. vulgaris cell wall was digested with enzyme mixture for preparing protoplasts. The optimal transformation efficiency was 1.67 × 104 ± 0.083 cfu μg−1 plasmid under the following conditions: 2 × 106 cells mL−1 of growing culture; 655 V pulse voltage with 3.4 ms pulse width. After transformation, green and cyan fluorescence were observed from transgenic C. vulgaris harboring gfp (green fluorescent protein) gene of pCAMBIA1302 and cfp (cyan fluorescent protein) gene of pSK397, respectively, using laser confocal microscope. RT-PCR analysis as well as Southern blot confirmed the integration of reporter gene at the molecular level. This efficient transformation system of C. vulgaris would be valuable for the production of recombinant proteins in the future.








Similar content being viewed by others
References
Bai LL, Yin WB, Chen YH, Niu LL, Sun YR, Zhao SM, Yang FQ, Wang RRC, Wu Q, Zhang XQ, Hu ZM (2013) A new strategy to produce a defensin: stable production of mutated NP-1 in nitrate reductase-deficient C. ellipsoidea. PLoS One 8:e54966. https://doi.org/10.1371/journal.pone.0054966
Berrios H, Zapata M, Rivas M (2016) A method for genetic transformation of Botryococcus braunii using a cellulase pretreatment. J Appl Phycol 28:201–208
Beyerinck [Beijerinck], MW (1890) Culturversuche mit Zoochlorellen, Lichenengonidien und anderen niederen Algen. Botanische Zeitung 48:725–785
Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, Salamov A, Terry A, Yamada T, Dunigan DD, Grigoriev IV, Claverie JM, van Etten JL (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22:2943–2955
Burlew JS (ed) (1953) Algal culture from laboratory to pilot plant. Carnegie Institution of Washington, Washington DC
Cha TS, Yee W, Aziz A (2012) Assessment of factors affecting Agrobacterium-mediated genetic transformation of the unicellular green alga, Chlorella vulgaris. World J Microbiol Biotechnol 28:1771–1779
Chatzikonstantinou M, Kalliampakou A, Gatzogia M, Flemetakis E, Katharios P, Labrou NE (2017) Comparative analyses and evaluation of the cosmeceutical potential of selected Chlorella strains. J Appl Phycol 29:179–188
Chien LF, Kuo TT, Liu BH, Lin HD, Feng TY, Huang CC (2012) Solar-to-bioH2 production enhanced by homologous overexpression of hydrogenase in green alga Chlorella sp. DT. Int J Hydrog Energy 37:17738–17748
Converti A, Casazza AA, Ortiz EY, Perego P, Del Borghi M (2009) Effect of temperature and nitrogen concentration on the growth and lipid content of Nannochloropsis oculata and Chlorella vulgaris for biodiesel production. Chem Eng Process Process Intensif 48:1146–1151
Dawson HN, Burlingame R, Cannons AC (1997) Stable transformation of Chlorella: rescue of nitrate reductase-deficient mutants with the nitrate reductase gene. Curr Microbiol 35:356–362
de Souza Queiroz J, Barbosa CM, da Rocha MC, Bincoletto C, Paredes-Gamero EJ, de Souza Queiroz ML, Palermo Neto J (2012) Chlorella vulgaris treatment ameliorates the suppressive effects of single and repeated stressors on hematopoiesis. Brain Behav Immun 29:39–50
Fan JH, Xu H, Luo YC, Wan MX, Huang JK, Wang WL, Li YG (2015a) Impacts of CO2 concentration on growth, lipid accumulation, and carbon-concentrating-mechanism related gene expression in oleaginous Chlorella. Appl Microbiol Biotechnol 99:2451–2462
Fan J, Ning K, Zeng X, Luo Y, Wang D, Hu J, Jing L, Hui X, Huang J, Wan M, Wang W, Zhang D, Shen G, Run C, Liao J, Fang L, Huang S, Jing X, Su X, Wang A, Bai L, Hu Z, Xu J, Liet Y (2015b) Genomic foundation of starch-to-lipid switch in oleaginous Chlorella spp. Plant Physiol 169:2444–2461
Gao C, Wang Y, Shen Y, Yan D, He X, Dai J, Wu Q (2014) Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genomics 15:582
Geng DG, Han Y, Wang YQ, Wang P, Zhang LM, Li WB, Sun YR (2004) Construction of a system for the stable expression of foreign genes in Dunaliella salina. Acta Bot Sinica 46:342–346
Gerken HG, Donohoe B, Knoshaug EP (2013) Enzymatic cell wall degradation of Chlorella vulgaris and other microalgae for biofuels production. Planta 237:239–253
Gong Y, Hu H, Gao Y, Xu X, Gao H (2011) Microalgae as platforms for production of recombinant proteins and valuable compounds: progress and prospects. J Ind Microbiol Biotechnol 38:1879–1890
Guo SL, Zhao XQ, Tang Y, Wan C, Alam MA, Ho SH, Bai FW, Chang JS (2013) Establishment of an efficient genetic transformation system in Scenedesmus obliquus. J Biotechnol 163:61–68
Hawkins RL, Nakamura M (1999) Expression of human growth hormone by the eukaryotic alga, Chlorella. Curr Microbiol 38:35–341
Jarvis EE, Brown LM (1991) Transient expression of firefly luciferase in protoplasts of the green alga Chlorella ellipsoidea. Curr Genet 19:317–321
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jeon K, Suresh A, Kim YC (2013) Highly efficient molecular delivery into Chlamydomonas reinhardtii by electroporation. Korean J Chem Eng 30:1626–1630
Jeong H, Lim JM, Park J, Sim Y, Choi HG, Lee J, Jeong WJ (2014) Plastid and mitochondrion genomic sequences from Arctic Chlorella sp. ArM0029B. BMC Genomics 15:286
Justo GZ, Silva MR, Queiroz ML (2001) Effects of the green algae Chlorella vulgaris on the response of the host hematopoietic system to intraperitoneal Ehrlich ascites tumor transplantation in mice. Immunopharmacol Immunotoxicol 23:119–132
Kilian O, Benemann CS, Niyogi KK, Vick B (2011) High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc Natl Acad Sci 108:21265–21269
Kim D, Kim YT, Cho JJ, Bae J, Hur S, Hwang I, Choi T (2002) Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar Biotechnol 4:63–73
Kindle KL (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A 87:1228–1232
Kitada K, Machmudah S, Sasaki M, Goto M, Nakashima Y, Kumamoto S, Hasegawa T (2009) Supercritical CO2 extraction of pigment components with pharmaceutical importance from Chlorella vulgaris. J Chem Technol Biotechnol 84:657–661
Koo J, Park D, Kim H (2013) Expression of bovine lactoferrin N-lobe by the green alga, Chlorella vulgaris. Algae 28:379–387
Konishi F, Tanaka K, Himeno K, Taniguchi K, Nomoto K (1985) Antitumor effect induced by a hot water extract of Chlorella vulgaris (CE): resistance to meth-a tumor growth mediated by CE-induced polymorphonuclear leukocytes. Cancer Immunol Immunother 19:73–78
Kothari R, Pathak VV, Kumar V, Singh DP (2012) Experimental study for growth potential of unicellular alga Chlorella pyrenoidosa on dairy waste water: an integrated approach for treatment and biofuel production. Bioresour Technol. 116:466–470
Kumar SV, Misquitta RW, Reddy VS, Rao BJ, Rajam MV (2004) Genetic transformation of the green alga Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci 166:731–738
Kumar M, Choi J, An G, Kim SR (2017) Ectopic expression of OsSta2 enhances salt stress tolerance in rice. Front Plant Sci 8:316
Lin HD, Liu BH, Kuo TT, Tsai HC, Feng TY, Huang CC, Chien LF (2013) Knockdown of PsbO leads to induction of HydA and production of photobiological H-2 in the green alga Chlorella sp. DT. Bioresour Technol 143:154–162
Liu L, Wang Y, Zhang Y, Chen X, Zhang P, Ma S (2013) Development of a new method for genetic transformation of the green alga Chlorella ellipsoidea. Mol Biotechnol 54:211–219
Liu Z, Zhang L, Pu Y, Liu Z, Li Z, Zhao Y, Qin S (2014) Cloning and expression of a cytosolic HSP90 gene in Chlorella vulgaris. Biomed Res Int2014:487050
Lou S, Wang L, He L, Wang Z, Wang G, Lin X (2016) Production of crocetin in transgenic Chlorella vulgaris expressing genes crtRB and ZCD1. J Appl Phycol 28:1657–1665
Morimoto T, Nagatsu A, Murakami N, Sakakibara J, Tokuda H, Nishino H, Iwashima A (1995) Anti-tumour-promoting glyceroglycolipids from the green alga, Chlorella vulgaris. Phytochemistry 40:1433–1437
Morris HJ, Carrillo OV, Almarales Á, Bermúdez RC, Alonso ME, Borges L, Quintana MM, Fontaine R, Llauradó G, Hernández M (2009) Protein hydrolysates from the alga Chlorella vulgaris 87/1 with potentialities in immunonutrition. Biotechnol Appl 26:16–15
Murray HG, Thompson WF (1980) Rapid isolation of high molecular weight DNA. Nucleic Acids Res 8:4321–4325
Nam K, Lee H, Heo S-W, Chang YK, Han J-I (2017) Cultivation of Chlorella vulgaris with swine wastewater and potential for algal biodiesel production. J Appl Phycol 29:1171–1178
Niu YF, Zhang MH, Xie WH, Li JN, Gao YF, Yang WD, Liu JS, Li HY (2011) A new inducible expression system in a transformed green alga, Chlorella vulgaris. Genet Mol Res 10:3427–3434
Phukan MM, Chutia RS, Konwar B, Kataki R (2011) Microalgae Chlorella as a potential bio-energy feedstock. Appl Energy 88:3307–3312
Popper ZA, Michel G, Hervé C, Domozych DS, Willats WGT, Tuohy MG, Kloareg B, Stengel DB (2011) Evolution and diversity of plant cell walls: from algae to flowering plants. Annu Rev Plant Biol 62:8.1–8.24
Ren XY, Wang HQ, Zhu JB, Kong QJ (2010) Selection of Chlorella transformed with rotavirus VP4-ST fusion gene. Vet. Sci China 40:41–44
Run C, Fan g L, Fan J, Fan C, Luo Y, Hu Z, Li Y (2016) Stable nuclear transformation of the industrial alga Chlorella pyrenoidosa. Algal Res 17:196–201
Safi C, Zebib B, Merah O, Pontalier PY, Vaca-Garcia C (2014) Morphology, composition, production, processing and applications of Chlorella vulgaris: a review renew. Sust Energ Rev 35:265–278
Siemering KR, Golbik R, Sever R, Haseloff J (1996) Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol 6:1653–1663
Singh A, Singh SP, Bamezai R (1999) Inhibitory potential of Chlorella vulgaris (E-25) on mouse skin papillomagenesis and xenobiotic detoxication system. Anticancer Res 19:1887–1891
Stanier RY, Kunisawa R, Mandel M, Cohen-Bazire G (1971) Purification and properties of unicellular blue-green algae (order Chroococcales). Bact Rev 35:171–205
Talebi AF, Tohidfar M, Tabatabaei M, Bagheri A, Mohsenpor M, Mohtashami SK (2013) Genetic manipulation, a feasible tool to enhance unique characteristic of Chlorella vulgaris as a feedstock for biodiesel production. Mol Biol Rep 40:4421–4428
Wang C, Wang Y, Su Q, Gao X (2007) Transient expression of GUS gene in a unicellular marine green alga, Chlorella sp. MACC/C95, via electroporation. Biotechnol Bioeng 12:180–183
Wang L, Chen X, Wang H, Zhang Y, Tang Q, Li J (2017a) Chlorella vulgaris cultivation in sludge extracts from 2, 4, 6-TCP wastewater treatment for toxicity removal and utilization. J Environ Manag 187:146–153
Wang P, Wong Y-S, Tam NF-Y (2017b) Green microalgae in removal and biotransformation of estradiol and ethinylestradiol. J Appl Phycol 29:263–273
Xue J, Wang L, Zhang L, Balamurugan S, Li D-W, Zeng H, Yang WD, Liu JS, Li HY (2016) The pivotal role of malic enzyme in enhancing oil accumulation in green microalga Chlorella pyrenoidosa. Microb Cell Factories 15:120
Yang B, Liu J, Liu B, Sun P, Ma X, Jiang Y, Wei D, Chen F (2015) Development of a stable genetic system for Chlorella vulgaris—a promising green alga for CO2 biomitigation. Algal Res 12:134–141
Yang B, Liu J, Jiang Y, Chen F (2016) Chlorella species as hosts for genetic engineering and expression of heterologous proteins: progress, challenge and perspective. Biotechnol J 11:1244–1261
Yamamoto M, Fujishita M, Hirata A, Kawano S (2004) Regeneration and maturation of daughter cell walls in the autospore-forming green alga Chlorella vulgaris (Chlorophyta, Trebouxiophyceae). J Plant Res 117:257–264
Yasukawa K, Akihisa T, Kanno H, Kaminaga T, Izumida M, Sakoh T, Tamura T, Takido M (1996) Inhibitory effects of sterols isolated from Chlorella vulgaris on 12-0-tetradecanoylphorbol-13-acetate-induced inflammation and tumor promotion in mouse skin. Biol Pharm Bull 19:573–576
Zheng H, Yin J, Gao Z, Huang H, Ji X, Dou C (2011) Disruption of Chlorella vulgaris cells for the release of biodiesel-producing lipids: a comparison of grinding, ultrasonication, bead milling, enzymatic lysis, and microwaves. Appl Biochem Biotechnol 164:1215–1224
Zuñiga C, Li CT, Huelsman T, Levering J, Zielinski DC, McConnell BO, Long CP, Knoshaug EP, Guarnieri MT, Antoniewicz MR (2016) Genome-scale metabolic model for the green alga Chlorella vulgaris UTEX 395 accurately predicts phenotypes under autotrophic, heterotrophic, and mixotrophic growth conditions. Plant Physiol 172:589–602
Acknowledgments
This research was supported by grants from the LINK program, Korean Research Foundation, and from PAP program (PE17900) of Korea Polar Research Institute, Republic of Korea.
Authors’ contributions.
SRK conceived and designed study. MK conducted overall experiments, drafted manuscript and SRK finalized the manuscript. JJ maintained and measured the growth curves and analyzed transformants. JC conducted transformants analysis at protein level. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Fig. S1
Vector map showing the cloning of CrCFP expression vector pSK397. (GIF 116 kb)
High Resolution Image
(TIFF 474 kb)
Fig. S2
Expression of hygromycin phosphotransferase gene was analyzed in pCAMBIA1302 transformant of C. vulgaris. (GIF 51 kb)
High Resolution Image
(TIFF 250 kb)
Rights and permissions
About this article
Cite this article
Kumar, M., Jeon, J., Choi, J. et al. Rapid and efficient genetic transformation of the green microalga Chlorella vulgaris. J Appl Phycol 30, 1735–1745 (2018). https://doi.org/10.1007/s10811-018-1396-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-018-1396-3