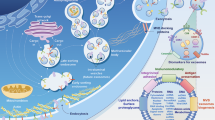
Abstract
Periodontitis is a prevalent and persistent inflammatory condition that impacts the supporting tissues of the teeth, including the gums and bone. Recent research indicates that mitochondrial dysfunction may be involved in the onset and advancement of periodontitis. The current work sought to reveal the interaction between mitochondrial dysfunction and the immune microenvironment in periodontitis. Public data were acquired from MitoCarta 3.0, Mitomap, and GEO databases. Hub markers were screened out by five integrated machine learning algorithms and verified by laboratory experiments. Single-cell sequencing data were utilized to unravel cell-type specific expression levels of hub genes. An artificial neural network model was constructed to discriminate periodontitis from healthy controls. An unsupervised consensus clustering algorithm revealed mitochondrial dysfunction-related periodontitis subtypes. The immune and mitochondrial characteristics were calculated using CIBERSORTx and ssGSEA algorithms. Two hub mitochondria-related markers (CYP24A1 and HINT3) were identified. Single-cell sequencing data revealed that HINT3 was primarily expressed in dendritic cells, while CYP24A1 was mainly expressed in monocytes. The hub genes based artificial neural network model showed robust diagnostic performance. The unsupervised consensus clustering algorithm revealed two distinct mitochondrial phenotypes. The hub genes exhibited a strong correlation with the immune cell infiltration and mitochondrial respiratory chain complexes. The study identified two hub markers that may serve as potential targets for immunotherapy and provided a novel reference for future investigations into the function of mitochondria in periodontitis.











Similar content being viewed by others
AVAILABILITY OF DATA AND MATERIAL
The date and materials of the current study are available from the corresponding author on reasonable request.
Abbreviations
- ROS:
-
Reactive oxygen species
- OXPHOS:
-
Oxidative phosphorylation
- mtDAMPs:
-
Mitochondrial damage-associated molecular patterns
- PRR:
-
Pattern recognition receptors
- UMAP:
-
Uniform manifold approximation and projection
- GEO:
-
Gene Expression Omnibus
- DEGs:
-
Differentially expressed genes
- FC:
-
Fold-change
- PCA:
-
Principal component analysis
- DMRGs:
-
Differentially expressed mitochondria-related genes
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LASSO:
-
Absolute shrinkage and selection operator
- SVM-RFE:
-
Support vector machine recursive feature elimination
- ANN:
-
Artificial neural network
- ROC:
-
Receiver operating characteristic
- DCA:
-
Decision curve analysis
- CIBERSORTx:
-
Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts x
- GSEA:
-
Gene set enrichment analysis
- ssGSEA:
-
Single-sample gene set enrichment analysis
- WGCNA:
-
Weighted correlation network analysis
- MM:
-
Module membership
- GS:
-
Gene significance
- CDF:
-
Cumulative distribution function
- AUC:
-
Area under the curve
- MitoPathway:
-
Mitochondria-related pathway
- CT:
-
Cycle threshold
- CtTp:
-
Ct of target genes for periodontitis sample
- CtGp:
-
Ct of house-keeping genes for periodontitis sample
- CtGc:
-
Ct of housekeeping gene for controls
- CtTc:
-
Ct of target genes for control
References
Pihlstrom, B.L., B.S. Michalowicz, and N.W. Johnson. 2005. Periodontal diseases. The Lancet 366 (9499): 1809–1820.
Hajishengallis, G. 2015. Periodontitis: From microbial immune subversion to systemic inflammation. Nature Reviews Immunology 15 (1): 30–44.
Eke, P.I., W.S. Borgnakke, and R.J. Genco. 2020. Recent epidemiologic trends in periodontitis in the USA. Periodontol 2000 82 (1): 257–267.
Zong, W.X., J.D. Rabinowitz, and E. White. 2016. Mitochondria and cancer. Molecular Cell 61 (5): 667–676.
Glancy, B. 2020. Visualizing mitochondrial form and function within the cell. Trends in Molecular Medicine 26 (1): 58–70.
Marchi, S., et al. 2022. Mitochondrial control of inflammation. Nature Reviews Immunology 1–15.
Liu, J., et al. 2018. P53 mediates lipopolysaccharide-induced inflammation in human gingival fibroblasts. Journal of Periodontology 89 (9): 1142–1151.
Liu, J., et al. 2022. Mitochondrial DNA efflux maintained in gingival fibroblasts of patients with periodontitis through ROS/mPTP pathway. Oxidative Medicine and Cellular Longevity 2022: 1000213.
Li, A., et al. 2022. Periodontitis and cognitive impairment in older adults: The mediating role of mitochondrial dysfunction. Journal of Periodontology.
Chen, J., et al. 2021. Sirtuin 3 deficiency exacerbates age-related periodontal disease. Journal of Periodontal Research 56 (6): 1163–1173.
Kam, A.Y.F., et al. 2021. Selective ERBB2 and BCL2 inhibition is synergistic for mitochondrial-mediated apoptosis in MDS and AML cells. Molecular Cancer Research 19 (5): 886–899.
Foo, J., et al. 2022. Mitochondria-mediated oxidative stress during viral infection. Trends in Microbiology 30 (7): 679–692.
Sun, H., et al. 2023. Melatonin promoted osteogenesis of human periodontal ligament cells by regulating mitochondrial functions through the translocase of the outer mitochondrial membrane 20. Journal of Periodontal Research 58 (1): 53–69.
Chen, M., et al. 2019. Oxidative stress-related biomarkers in saliva and gingival crevicular fluid associated with chronic periodontitis: A systematic review and meta-analysis. Journal of Clinical Periodontology 46 (6): 608–622.
Hyeon, S., et al. 2013. Nrf2 deficiency induces oxidative stress and promotes RANKL-induced osteoclast differentiation. Free Radical Biology & Medicine 65: 789–799.
Liu, J., et al. 2022. Abnormal mitochondrial structure and function are retained in gingival tissues and human gingival fibroblasts from patients with chronic periodontitis. Journal of Periodontal Research 57 (1): 94–103.
Darveau, R.P. 2010. Periodontitis: A polymicrobial disruption of host homeostasis. Nature Reviews Microbiology 8 (7): 481–490.
Shen, K.L., et al. 2022. Effects of artificial intelligence-assisted dental monitoring intervention in patients with periodontitis: A randomized controlled trial. Journal of Clinical Periodontology 49 (10): 988–998.
Vollmer, A., et al. 2022. Associations between periodontitis and COPD: An artificial intelligence-based analysis of NHANES III. Journal of Clinical Medicine 11(23).
Wang, Z., et al. 2023. DNER and GNL2 are differentially m6A methylated in periodontitis in comparison with periodontal health revealed by m6A microarray of human gingival tissue and transcriptomic analysis. Journal of Periodontal Research 58 (3): 529–543.
Kim, H., et al. 2021. Differential DNA methylation and mRNA transcription in gingival tissues in periodontal health and disease. Journal of Clinical Periodontology 48 (9): 1152–1164.
Peng, L., et al. 2022. Identification and validation of a classifier based on hub aging-related genes and aging subtypes correlation with immune microenvironment for periodontitis. Frontiers in Immunology 13: 1042484.
Chen, H., et al. 2022. Exploration of cross-talk and pyroptosis-related gene signatures and molecular mechanisms between periodontitis and diabetes mellitus via peripheral blood mononuclear cell microarray data analysis. Cytokine 159: 156014.
Williams, D.W., et al. 2021. Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell 184 (15): 4090-4104.e15.
Caetano, A.J., et al. 2021. Defining human mesenchymal and epithelial heterogeneity in response to oral inflammatory disease. Elife 10.
Caetano, A.J., et al. 2023. Spatially resolved transcriptomics reveals pro-inflammatory fibroblast involved in lymphocyte recruitment through CXCL8 and CXCL10. Elife 12.
Lundmark, A., et al. 2018. Gene expression profiling of periodontitis-affected gingival tissue by spatial transcriptomics. Science and Reports 8 (1): 9370.
Tonetti, M.S., H. Greenwell, and K.S. Kornman. 2018. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. Journal of Periodontology 89 (Suppl 1): S159-s172.
Caton, J.G., et al. 2018. A new classification scheme for periodontal and peri-implant diseases and conditions - Introduction and key changes from the 1999 classification. Journal of Clinical Periodontology 45 (Suppl 20): S1–s8.
Kowaltowski, A.J., A.E. Vercesi, and G. Fiskum. 2000. Bcl-2 prevents mitochondrial permeability transition and cytochrome c release via maintenance of reduced pyridine nucleotides. Cell Death and Differentiation 7 (10): 903–910.
Chen, H., et al. 2022. Pyroptosis may play a crucial role in modifications of the immune microenvironment in periodontitis. Journal of Periodontal Research 57 (5): 977–990.
Kebschull, M., et al. 2014. Gingival tissue transcriptomes identify distinct periodontitis phenotypes. Journal of Dental Research 93 (5): 459–468.
Abe, D., et al. 2011. Altered gene expression in leukocyte transendothelial migration and cell communication pathways in periodontitis-affected gingival tissues. Journal of Periodontal Research 46 (3): 345–353.
Demmer, R.T., et al. 2008. Transcriptomes in healthy and diseased gingival tissues. Journal of Periodontology 79 (11): 2112–2124.
Liu, Y., et al. 2020. Long non-coding RNA and mRNA expression profiles in peri-implantitis vs periodontitis. Journal of Periodontal Research 55 (3): 342–353.
Taminau, J., et al. 2012. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinformatics 13 (1): 335.
Chen, C., et al. 2011. Removing batch effects in analysis of expression microarray data: An evaluation of six batch adjustment methods. PLoS ONE 6 (2): e17238.
Stuart, T., et al. 2019. Comprehensive Integration of Single-Cell Data. Cell 177 (7): 1888–1902.e21.
Aran, D., A.P. Looney, L. Liu, E. Wu, V. Fong, A. Hsu, S. Chak, R.P. Naikawadi, P.J. Wolters, A.R. Abate, A.J. Butte, and M. Bhattacharya. 2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nature Immunology 20 (2): 163–172. https://doi.org/10.1038/s41590-018-0276-y. Epub 2019 Jan 14. PMID: 30643263; PMCID: PMC6340744.
Monaco, G., et al. 2019. RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Reports 26 (6): 1627-1640.e7.
Rath, S., et al. 2021. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Research 49 (D1): D1541–d1547.
Shen, W., et al. 2022. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. Imeta 1 (3): e36.
Tolles, J., and W.J. Meurer. 2016. Logistic regression: Relating patient characteristics to outcomes. JAMA 316 (5): 533–534.
Hänzelmann, S., R. Castelo, and J. Guinney. 2013. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14: 7.
Sanz, H., et al. 2018. SVM-RFE: Selection and visualization of the most relevant features through non-linear kernels. BMC Bioinformatics 19 (1): 432.
Li, W., et al. 2019. Gene expression value prediction based on XGBoost algorithm. Frontiers in Genetics 10: 1077.
Chen, X., and H. Ishwaran. 2012. Random forests for genomic data analysis. Genomics 99 (6): 323–329.
Beck, M.W. 2018. NeuralNetTools: Visualization and analysis tools for neural networks. Journal of Statistical Software 85 (11): 1–20.
Nachid, M., and M. Boussiala. 2021. Machine Learning_Iris_caret~Package.
Vickers, A.J., and E.B. Elkin. 2006. Decision curve analysis: A novel method for evaluating prediction models. Medical Decision Making 26 (6): 565–574.
Newman, A.M., et al. 2019. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nature Biotechnology 37 (7): 773–782.
Yu, G., et al. 2012. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology 16 (5): 284–287.
Singh, L.N., et al. 2021. MitoScape: A big-data, machine-learning platform for obtaining mitochondrial DNA from next-generation sequencing data. PLoS Computational Biology 17 (11): e1009594.
Langfelder, P., and S. Horvath. 2008. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559.
Wilkerson, M.D., and D.N. Hayes. 2010. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 26 (12): 1572–1573.
Zhang, B., et al. 2020. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Molecular Cancer 19 (1): 53.
Chan-Seng-Yue, M., et al. 2020. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nature Genetics 52 (2): 231–240.
Zindel, J., and P. Kubes. 2020. DAMPs, PAMPs, and LAMPs in Immunity and Sterile Inflammation. Annual Review of Pathology: Mechanisms of Disease 15: 493–518.
Grazioli, S., and J. Pugin. 2018. Mitochondrial damage-associated molecular patterns: From inflammatory signaling to human diseases. Frontiers in Immunology 9: 832.
Lin, M.M., et al. 2022. Mitochondrial-derived damage-associated molecular patterns amplify neuroinflammation in neurodegenerative diseases. Acta Pharmacologica Sinica.
Scazzone, C., et al. 2021. Vitamin D and Genetic Susceptibility to Multiple Sclerosis. Biochemical Genetics 59 (1): 1–30.
Tuckey, R.C., et al. 2023. Analysis of the ability of vitamin D3-metabolizing cytochromes P450 to act on vitamin D3 sulfate and 25-hydroxyvitamin D3 3-sulfate. Journal of Steroid Biochemistry and Molecular Biology 227: 106229.
Al-Zahrani, M.S. 2006. Increased intake of dairy products is related to lower periodontitis prevalence. Journal of Periodontology 77 (2): 289–294.
Zihni Korkmaz, M., et al. 2022. The effects of vitamin D deficiency on mandibular bone structure: a retrospective radiological study. Oral Radiology 1–8.
Hu, Z., F. Zhou, and H. Xu. 2022. Circulating vitamin C and D concentrations and risk of dental caries and periodontitis: A Mendelian randomization study. Journal of Clinical Periodontology 49 (4): 335–344.
Shahijanian, F., et al. 2014. The CYP27B1 variant associated with an increased risk of autoimmune disease is underexpressed in tolerizing dendritic cells. Human Molecular Genetics 23 (6): 1425–1434.
Kundu, R., et al. 2014. Regulation of CYP27B1 and CYP24A1 hydroxylases limits cell-autonomous activation of vitamin D in dendritic cells. European Journal of Immunology 44 (6): 1781–1790.
Brenner, C. 2002. Hint, Fhit, and GalT: Function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry 41 (29): 9003–9014.
Wang, W., et al. 2013. All-trans retinoic acid protects hepatocellular carcinoma cells against serum-starvation-induced cell death by upregulating collagen 8A2. FEBS Journal 280 (5): 1308–1319.
Derakhshani, A., et al. 2021. The role of hemoglobin subunit delta in the immunopathy of multiple sclerosis: Mitochondria matters. Frontiers in Immunology 12: 709173.
Vercellino, I., and L.A. Sazanov. 2022. The assembly, regulation and function of the mitochondrial respiratory chain. Nature Reviews Molecular Cell Biology 23 (2): 141–161.
Ledderose, C., et al. 2015. Novel method for real-time monitoring of ATP release reveals multiple phases of autocrine purinergic signalling during immune cell activation. Acta Psychologica 213 (2): 334–345.
González-Arzola, K., et al. 2019. New moonlighting functions of mitochondrial cytochrome c in the cytoplasm and nucleus. FEBS Letters 593 (22): 3101–3119.
Santucci, R., et al. 2019. Cytochrome c: An extreme multifunctional protein with a key role in cell fate. International Journal of Biological Macromolecules 136: 1237–1246.
Barrera, M.J., et al. 2021. Dysfunctional mitochondria as critical players in the inflammation of autoimmune diseases: Potential role in Sjögren’s syndrome. Autoimmunity Reviews 20 (8): 102867.
Gogvadze, V., S. Orrenius, and B. Zhivotovsky. 2006. Multiple pathways of cytochrome c release from mitochondria in apoptosis. Biochimica et Biophysica Acta 1757 (5–6): 639–647.
Jiang, X., and X. Wang. 2004. Cytochrome C-mediated apoptosis. Annual Review of Biochemistry 73: 87–106.
Liu, Q., et al. 2022. Inhibition of TRPA1 ameliorates periodontitis by reducing periodontal ligament cell oxidative stress and apoptosis via PERK/eIF2α/ATF-4/CHOP signal pathway. Oxidative Medicine and Cellular Longevity 2022: 4107915.
Lucas, H., et al. 2010. Inhibition of apoptosis in periodontitis. Journal of Dental Research 89 (1): 29–33.
Quiles, J.M., and B. Gustafsson Å. 2022. The role of mitochondrial fission in cardiovascular health and disease. Nature Reviews Cardiology.
Gillette, M.A., et al. 2020. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell 182 (1): 200-225.e35.
Kovalev, E.V., and S.A. Gusev. 1988. Age-related changes in the ultrastructural organization of the human gingival epithelium. Arkhiv Anatomii, Gistologii i Émbriologii 94 (4): 44–47.
Varela-Lopez, A., et al. 2016. Coenzyme Q protects against age-related alveolar bone loss associated to n-6 polyunsaturated fatty acid rich-diets by modulating mitochondrial mechanisms. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences 71 (5): 593–600.
ACKNOWLEDGEMENTS
We would like to exert a compelling appreciation for the GEO projects. We thank Dr. Jianming Zeng (University of Macau), Lao Junjun, Biomamba, and all members of their bioinformatics team, biotrainee, for generously sharing their experience and codes.
Funding
This work was financially supported by the National Natural Science Foundation of China [grant number 81700982]; the Chongqing Medical Reserve Talent Studio for Young People [grant number ZQNYXGDRCGZS2019004].
Author information
Authors and Affiliations
Contributions
Xiaonan Zhang: supervision, project administration, funding acquisition. Hang Chen: conceptualization, methodology, software, investigation, formal analysis, visualization, validation, writing—original draft. Limin Peng: methodology, visualization, validation, resources, writing—review and editing. Zhenxiang Wang: data curation, validation, writing—review and editing. Yujuan He: writing—review and editing.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
This study was conducted in accordance with the principles outlined in the Declaration of Helsinki and was approved by the Ethics Committee of the Stomatological Hospital of Chongqing Medical University (Approval No: 2020 LSNo.79).
Consent for Publication
All authors have approved the manuscript for submission.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10753_2023_1851_MOESM1_ESM.pdf
Supplementary Table 1: The detail of clinical information on 24 patients. Supplementary Table 2: Inclusion criteria and exclusion criteria for clinical participants. Supplementary Table 3: Characteristics of the bulk tissue transcriptomic datasets included in the study. Supplementary Table 4: Characteristics of the single-cell transcriptomic datasets included in the study. Supplementary Table 5: The primer sequences for qRT-PCR. Supplementary Table 6: The output results of the neural network model. Supplementary Fig. 1: The WGCNA results. (PDF 365 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, H., Peng, L., Wang, Z. et al. Integrated Machine Learning and Bioinformatic Analyses Constructed a Network Between Mitochondrial Dysfunction and Immune Microenvironment of Periodontitis. Inflammation 46, 1932–1951 (2023). https://doi.org/10.1007/s10753-023-01851-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-023-01851-0