Abstract
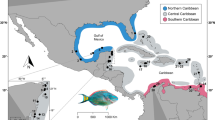
An increasing number of studies are applying novel genetic tools to assess familial relationships of marine organisms to underpin management and conservation efforts. Here, we describe the reproductive population structure of the endangered Atlantic Chupare stingray (Styracura schmardae) from The Bahamas using a double digest restriction-site-associated DNA methodology among 71 individuals. Results from the 1552 SNPs indicate clear kinship patterns in 52 stingrays collected from three geographic zones and suggest individuals from the Exuma Sound study area belong to a single genetic population. A gradient in mature-immature rays existed, suggesting the Chupare rays of Exuma Sound may be reliant on the shallow waters along this island chain as an ontogenetic migration corridor (the deep waters of the sound likely is a physical barrier). These data are critical in evaluating the importance of coastal shallow water habitats to population connectivity in this large coastal elasmobranch. Study results document inherent vulnerability within the Exuma Sound population. Further, these data provide additional support for the declining population trends noted across the range of this endangered species and can inform effective management of this stingray and the habitats that support it.




Similar content being viewed by others
Data availability
The data that support this study will be shared upon reasonable request to the authors. Demultiplexed sequence data are available at the SRA database (SRA Bioproject: PRJNA905404).
Code availability
Code will be shared upon reasonable request to the authors.
References
Andrews, S., 2010. FastQC, A Quality Control Tool for High Throughput Sequence Data (Version 0.11.9). Available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Accessed January 2021].
Carvalho, G. R. & L. Hauser, 1995. Genetic impacts of fish introductions: a perspective on African lakes. In Pitcher, T. J. & P. B. J. Hart (eds), The Impact of Species Changes in African Lakes. Chapman and Hall Fish and Fisheries Series, Vol. 18. Springer, Dordecht: 457–493.
Catchen, J., P. A. Hohenlohe, S. Bassham, A. Amores & W. A. Cresko, 2013. Stacks: an analysis tool set for population genomics. Molecular Ecology 22: 3124–3140.
Danecek, P., A. Auton, G. Abecasis, C. A. Albers, E. Banks, M. A. DePristo, R. E. Handsaker, G. Lunter, G. T. Marth, S. T. Sherry, G. McVean & R. Durbin, 2011. 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics 27: 2156–2158.
Do, C., R. S. Waples, D. Peel, G. M. Macbeth, B. J. Tillet & J. R. Ovenden, 2014. NeEstimator V2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources 14(1): 209–214.
Dulvy, N. K., S. L. Fowler, J. A. Musick, R. D. Cavanagh, P. M. Kyne, L. R. Harrison, J. K. Carlson, L. N. K. Davidson, S. V. Fordham, M. P. Francis, C. M. Pollock, C. A. Simpfendorfer, G. H. Burgess, K. E. Carpenter, L. J. V. Compagno, D. A. Ebert, C. Gibson, M. R. Heupel, S. R. Livingstone, J. C. Saciango, J. D. Stevens, S. Valenti & W. White, 2014. Extinction risk and conservation of the world’s sharks and rays. Elife 3: e00590.
Dulvy, N. K., C. A. Simpfendorfer, L. N. Davidson, S. V. Fordham, A. Bräutigam, G. Sant & D. J. Welch, 2017. Challenges and priorities in shark and ray conservation. Current Biology 27: R565–R572.
Dulvy, N.K., P. Charvet, R. Pollom, C. Avalos, M.P. Blanco-Parra, A. Briones Bell-Iloch, D. Cardenosa, D. Derrick, E. Espinoza, P.A. Mejía-Falla, J.M. Morales-Saldaña, B. Naranjo-Elizondo, A.F. Navia, N.J. Simpson, & E.V.C. Schneider, 2021. Struacura schmardae. The IUCN Red List of Threatened Species 2021. https://www.iucnredlist.org/species/60161/3090840.
Earl, D. A. & B. M. von Holdt, 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361.
Evanno, G., S. Regnaut & J. Goudet, 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14(8): 2611–2620.
Faircloth, B., & T. Glenn, 2011. Homemade AMPure XP Beads. Department of Ecology and Evolutionary Biology. University of California, Los Angeles.
Gosselin, T., & E. Archer, 2019. Grur: an R package tailored for RADseq data imputations. R package version 0.1.4. https://github.com/thierrygosselin/grur.
Hickey, B. M., P. MacCready, E. Elliott & N. B. Kachel, 2000. Dense saline plumes in Exuma sound, Bahamas. Journal of Geophysical Research Oceans 105: 11471–11488.
Huisman, J., 2017. Pedigree reconstruction from SNP data: parentage assignment, sibship clustering and beyond. Molecular Ecology Resources. 17: 1009–1024.
Jakobsson, M. & N. A. Rosenberg, 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806.
Johri, S., M. P. Doane, L. Allen & E. A. Dinsdale, 2019. Taking advantage of the genomics revolution for monitoring and conservation of chondrichthyan populations. Diversity 11: 49.
Jombart, T. & I. Ahmed, 2011. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071.
Kamvar, Z. N., J. F. Tabima & N. J. Grünwald, 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2: e281.
Keenan, K., P. McGinnity, T. F. Cross, W. W. Crozier & P. A. Prodohl, 2013. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods in Ecology and Evolution 14: 782–788.
Kyne, P. M. & C. A. Simpfendorfer, 2010. Sharks and Their Relatives II: Biodiversity, Adaptive Physiology, and Conservation, 1st ed. CRC Press, Boca Raton.
Larson, W. A., D. A. Isermann & Z. S. Feiner, 2020. Incomplete bioninformatic filtering and inadequate age and growth analysis lead to an incorrect inference of harvested-induced changes. Evolutionary Applications 14: 278–289.
Last, P. R., J. D. Stevens, R. Swainston & G. Davis, 2009. Sharks and Rays of Australia, CSIRO Australia, Clayton.
Last, P., G. Naylor, B. Séret, W. White, M. de Carvalho & M. Stehmann, 2016. Rays of the World, CSIRO Publishing, Clayton.
Luikart, G., T. Antao, B. K. Hand, C. C. Muhlfeld, M. C. Boyer, T. Cosart, B. Trethewey, R. Al-Chockhachy & R. S. Waples, 2021. Detecting population declines via monitoring the effective number of breeders (Nb). Molecular Ecology Resources 21(2): 379–393.
Mastretta-Yanes, A., N. Arrigo, N. Alvarez, T. H. Jorgensen, D. Piñeros & B. C. Emerson, 2015. Restriction site-associated DNA sequencing, genotyping error estimation, and de novo assembly optimization for population genetic inference. Molecular Ecology Resources 15: 28–41.
McKinney, G. J., R. K. Waples, L. W. Seeb & J. E. Seeb, 2017. Paralogs are revealed by proportion of heterozygotes and deviations in read ratios in genotyping-by-sequencing data from natural populations. Molecular Ecology Resources 17: 656–669.
Mills, L. S. & F. W. Allendorf, 1996. The one-migrant-per-generation rule in conservation and management. Conservation Biology 10: 1509–1518.
Nunes, A. R. & J. L. S. Nunes, 2020. The mystery of Styracura schmardae stingrays from the Brazilian Amazon coast. Examines Marine Biology and Oceanography. https://doi.org/10.31031/EIMBO.2020.03.000564.
O’Shea, O. R., C. Ward & E. Brooks, 2017. Range Extension in Styracura (= Himantura) schmardae (Caribbean Whiptail Stingray) from The Bahamas. Caribbean Naturalist 38: 1–8.
O’Shea, O. R., M. H. Meadows, E. E. Wrigglesworth, J. Newton & L. A. Hawkes, 2020. Novel insights into the diet of southern stingrays and Caribbean whiptail rays. Marine Ecology Progress Series 655: 157–170.
O’Shea, O. R., T. E. Van Leeuwen, D. O. O’Brien, L. Arrowsmith, R. McCalman, M. Griffiths & D. A. Exton, 2021. Evidence and description of a nursery habitat for the recently reclassified stingray Styracura schmardae from The Bahamas. Marine Ecology Progress Series 660: 141–151.
Peterson, B. K., J. N. Weber, E. H. Kay, H. S. Fisher & H. E. Hoekstra, 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7: e37135.
Plank, S. M., C. G. Lowe, K. A. Feldheim, R. R. Wilson Jr. & J. A. Brusslan, 2010. Population genetic structure of the round stingray Urobatis helleri (Elasmobranchii: Rajiformes) in southern California and the Gulf of California. Journal of Fish Biology 77(2): 329–340.
Pritchard, J. K., M. Stephens & P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Raj, A., M. Stephens & J. K. Pritchard, 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197: 573–589.
Reid-Anderson, S., K. Bilgmann & A. Stow, 2019. Effective population size of the critically endangered east Australian grey nurse shark Carcarias taurus. Marine Ecology Progress Series 610: 137–148.
Richards, V. P., M. B. DeBiasse & M. Shivji, 2018. Deep mitochondrial lineage divergence among populations of the southern stingray (Hypanus americanus (Hildebrand & Schroeder, 1928)) throughout the southeastern United States and Caribbean. Marine Biodiversity 49(4): 1627–1634.
Rilov, G., A. D. Mazaris, V. Stelzenmüller, B. Helmuth, M. Wahl, T. Guy-Haim, N. Mieszkowska, J. B. Ledoux & S. Katsanevakis, 2019. Adaptive marine conservation planning in the face of climate change: what can we learn from physiological, ecological and genetic studies? Global Ecology and Conservation 17: e00566.
Sales, M. A. N., J. E. P. D. Freitas, C. C. Cavalcante, J. Santander-Neto, P. Charvet & V. V. Faria, 2020. The southernmost record and an update of the geographical range of the Atlantic chupare, Styracura schmardae (Chondrichthyes: Myliobatiformes). Journal of Fish Biology 97: 302–308.
Sandoval-Castillo, J., 2019. Conservation genetics of elasmobranchs of the Mexican Pacific Coast, trends and perspectives. Advances in Marine Biology 83: 115–157.
Schwanck, T. N., M. Schweinsberg, K. P. Lampert, T. L. Guttridge, R. Tollrian & O. R. O’Shea, 2020. Linking local movement and molecular analysis to explore philopatry and population connectivity of the southern stingray Hypanus americanus. Journal of Fish Biology 96: 1475–2148.
Shipley, O. N., K. J. Murchie, M. G. Frisk, E. J. Brooks, O. R. O’Shea & M. Power, 2017. Low polar compound effects and inter-tissue comparisons of stable isotope signatures in three nearshore elasmobranchs. Marine Ecology Progress Series 579: 233–238.
Shipley, O. N., K. J. Murchie, M. G. Frisk, O. R. O’Shea, M. Winchester, E. J. Brooks, J. Pearson & M. Power, 2018. Trophic niche dynamics of three nearshore benthic predators in The Bahamas. Hydrobiologia 813: 177–188.
Tan, M. P., L. L. Wong, S. A. Razali, N. Afiqah-Aleng, S. A. M. Nor, Y. Y. Sung, Y. Van de Peer, P. Soregloos & M. Danish-Daniel, 2019. Applications of next-generation sequencing technologies and computational tools in molecular evolution and aquatic animals conservation studies: a short review. Bioinformatics 15: 1176934319892284.
Wang, J., 2007. Triadic IBD coefficients and applications to estimating pairwise relatedness. Genetics Research 89(3): 135–153.
Wang, J., 2011. COANCESTRY: a program for simulating, estimating, and analysing relatedness and inbreeding coefficients. Molecular Ecology Resources 11(1): 141–145.
Waples, R. S., T. Antao & G. Luikart, 2014. Effects of overlapping generations on linkage disequilibrium estimates of effective population size. Genetics 197: 769–780.
Waples, R. K., W. A. Larson & R. S. Waples, 2016. Estimating contemporary effective population size in non-model species using linkage disequilibrium across thousands of loci. Heredity 117: 233–240.
Winter, D. J., 2012. MMOD: an R library for the calculation of population differentiation statistics. Molecular Ecology Resources 12(6): 1158–1160.
Acknowledgements
We thank the many volunteers and Cape Eleuthera Institute interns and technicians for assistance in data collection. A special thanks to Catherine Booker of the Exuma Foundation for in-kind support and for facilitating the outreach components of this study in Great Exuma. Additionally, A. Shultz, C. White, C. Ward, A. Waldman, K. Luniewicz, M. Firing, S. Burnside, D. Montgomery and R. Halinan for extended periods of field work. We also thank two anonymous reviewers for helpful suggestions which improved this manuscript.
Funding
This study was funded in part by the Hummingbird Cay Foundation, the Rufford Foundation and the Cape Eleuthera Foundation.
Author information
Authors and Affiliations
Contributions
Both authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by EW and ORO. The first draft of the manuscript was written by EW and ORO and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical approval
This study was conducted with permission from The Bahamas Department of Marine Resources Permit Nos. MAF/FIS/17 and MAF/FIS/34 and strictly followed IACUC regulations for animal care. It should be noted at the time of data collection, this species was considered ‘Data Deficient’ by the IUCN and so endangered species research permits were not required.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Handling editor: Cécile Fauvelot
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wallace, E.M., O’Shea, O.R. Population genetic connectivity of an endangered stingray from The Bahamas. Hydrobiologia 850, 441–454 (2023). https://doi.org/10.1007/s10750-022-05087-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-022-05087-1