Abstract
Background
Autophagy plays multifaceted roles in regulating hepatocellular carcinoma (HCC) and the mechanisms involved are under-explored. Regulatory microRNAs (miRNAs) have been reported to target autophagy proteins but their roles in HCC is not well studied. Using HCC patient tissues, this study aims to investigate the association of autophagy with several clinicopathological parameters as well as identifying the autophagy-related miRNAs and the possible pathways.
Methods and results
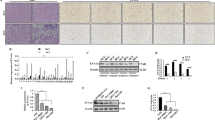
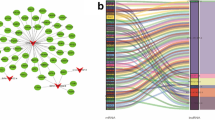
Autophagy level in the HCC patient-derived cancer and non-cancer tissues was determined by immunohistochemistry (IHC) targeting SQSTM1, LC3A and LC3B proteins. Significance tests of clinicopathological variables were tested using the Fisher’s exact or Chi-square tests. Gene and miRNA expression assays were carried out and analyzed using Nanostring platform and software followed by validation of other online bioinformatics tools, namely String and miRabel. Autophagy expression was significantly higher in cancerous tissues compared to adjacent non-cancer tissues. High LC3B expression was associated with advanced tumor histology grade and tumor location. Nanostring gene expression analysis revealed that SQSTM1, PARP1 and ATG9A genes were upregulated in HCC tissues compared to non-cancer tissues while SIRT1 gene was downregulated. These genes are closely related to an autophagy pathway in HCC. Further, using miRabel tool, three downregulated miRNAs (hsa-miR-16b-5p, hsa-miR-34a-5p, and hsa-miR-660-5p) and one upregulated miRNA (hsa-miR-539-5p) were found to closely interact with the abovementioned autophagy-related genes. We then mapped out the possible pathway involving the genes and miRNAs in HCC tissues.
Conclusions
We conclude that autophagy events are more active in HCC tissues compared to the adjacent non-cancer tissues. We also reported the possible role of several miRNAs in regulating autophagy-related genes in the autophagy pathway in HCC. This may contribute to the development of potential therapeutic targets for improving HCC therapy. Future investigations are warranted to validate the target genes reported in this study using a larger sample size and more targeted molecular technique.




Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Abdel-Moety A, Baddour N, Salem P et al (2022) SQSTM1 expression in hepatocellular carcinoma and relation to tumor recurrence after radiofrequency ablation. J Clin Exp Hepatol 12(3):774–784. https://doi.org/10.1016/j.jceh.2021.12.001
Akkoc Y, Gozuacik D (2018) Autophagy and liver cancer. Turk J Gastroenterol 29(3):270–282. https://doi.org/10.5152/tjg.2018.150318
Awi NJ, Armon S, Peh KB, Peh SC, Teow SY (2020) High expression of LC3A, LC3B, and p62/SQSTM1 autophagic proteins in human colonic ganglion cells. Malays J Pathol 42(1):85–90
Awi NJ, Yap HY, Armon S et al (2021) Association between autophagy and KRAS mutation with clinicopathological variables in colorectal cancer patients. Malays J Pathol 43(2):269–279
Chan HY, Ramasamy TS, Chung FF, Teow SY (2024) Role of sirtuin 1 (SIRT1) in regulation of autophagy and nuclear factor-kappa Beta (NF-ĸβ) pathways in sorafenib-resistant hepatocellular carcinoma (HCC). Cell Biochem Biophys. Published online March 11, 2024. https://doi.org/10.1007/s12013-024-01247-3
Chava S, Lee C, Aydin Y et al (2017) Chaperone-mediated autophagy compensates for impaired macroautophagy in the cirrhotic liver to promote hepatocellular carcinoma. Oncotarget 8(25):40019–40036. https://doi.org/10.18632/oncotarget.16685
Cicchini M, Karantza V, Xia B (2015) Molecular pathways: Autophagy in cancer- a matter of timing and context. Clin Cancer Res 21(3):498–504. https://doi.org/10.1158/1078-0432.CCR-13-2438
Dash S, Chava S, Chandra PK, Aydin Y, Balart LA, Wu T (2016) Autophagy in hepatocellular carcinomas: from pathophysiology to therapeutic response. Hepat Med 8:9–20. https://doi.org/10.2147/HMER.S63700
Gozuacik D, Akkoc Y, Ozturk DG, Kocak M (2017) Autophagy-regulating microRNAs and cancer. Front Oncol 7:65. https://doi.org/10.3389/fonc.2017.00065
Gu H, Gu S, Zhang X et al (2019) miR-106b-5p promotes aggressive progression of hepatocellular carcinoma via targeting RUNX3. Cancer Med 8(15):6756–6767. https://doi.org/10.1002/cam4.2511
Guardia CM, Tan XF, Lian T (2020) Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep 31(13):107837. https://doi.org/10.1016/j.celrep.2020.107837
Hao C, Zhu PX, Yang X et al (2014) Overexpression of SIRT1 promotes metastasis through epithelial-mesenchymal transition in hepatocellular carcinoma. BMC Cancer 14:978. https://doi.org/10.1186/1471-2407-14-978
Hennig P, Fenini G, Di Filippo M et al (2021) The pathways underlying the multiple roles of p62 in inflammation and cancer. Biomedicines 9(7). https://doi.org/10.3390/biomedicines9070707
Kessler SM, Pokorny J, Zimmer V et al (2013) IGF2 mRNA binding protein p62/IMP2-2 in hepatocellular carcinoma: antiapoptotic action is independent of IGF2/PI3K signaling. Am J Physiol Gastrointest Liver Physiol 304(4):G328–G336. https://doi.org/10.1152/ajpgi.00005.2012
Kimkong I, Kunanopparat A (2020) Autophagy related protein 9A increase in hepatitis B virus-associated hepatocellular carcinoma and the role in apoptosis. World J Hepatol 12(12):1367–1371. https://doi.org/10.4254/wjh.v12.i12.1367
Kobayashi M, Yasukawa H, Arikawa T et al (2021) Trehalose induces SQSTM1/p62 expression and enhances lysosomal activity and antioxidative capacity in adipocytes. FEBS Open Bio 11(1):185–194. https://doi.org/10.1002/2211-5463.13055
Li Y, Xu S, Li J et al (2016) SIRT1 facilitates hepatocellular carcinoma metastasis by promoting PGC-1α-mediated mitochondrial biogenesis. Oncotarget 7(20):29255–29274. https://doi.org/10.18632/oncotarget.8711
Li X, He S, Ma B (2020) Autophagy and autophagy-related proteins in cancer. Mol Cancer 19(1):12. https://doi.org/10.1186/s12943-020-1138-4
Liu L, Liao J, He X, Li P (2017) The role of autophagy in hepatocellular carcinoma: friend or foe. Oncotarget 8(34):57707–57722. https://doi.org/10.18632/oncotarget.17202
Liu YM, Cao Y, Zhao PS et al (2022) CircCCNB1 silencing acting as a miR-106b-5p sponge inhibited GPM6A expression to promote HCC progression by enhancing DYNC1I1 expression and activating the AKT/ERK signaling pathway: Erratum. Int J Biol Sci 18(6):2652–2654. https://doi.org/10.7150/ijbs.72651
Liu S, Zhang H, Yan J et al (2023) FOXP3 and SQSTM1/P62 correlate with prognosis and immune infiltration in hepatocellular carcinoma. Pathol Res Prac 242:154292. https://doi.org/10.1016/j.prp.2022.154292
Llovet JM, Kelley RK, Villanueva A et al (2021) Hepatocellular carcinoma. Nat Rev Dis Primers 7:6. https://doi.org/10.1038/s41572-020-00240-3
Luna A, Aladjem MI, Kohn KW (2013) SIRT1/PARP1 crosstalk: connecting DNA damage and metabolism. Genome Integr 4(1):6. https://doi.org/10.1186/2041-9414-4-6
Manley S, Williams JA, Ding WX (2013) Role of p62/SQSTM1 in liver physiology and pathogenesis. Exp Biol Med 238(5):525–538. https://doi.org/10.1177/1535370213489446
Meng YC, Lou XL, Yang LY et al (2020) Role of the autophagy-related marker LC3 expression in hepatocellular carcinoma: a metaanalysis. J Cancer Res Clin Oncol 146(5):1103–1113. https://doi.org/10.1007/s00432-020-03174-1
Morishita A, Oura K, Tadokoro T, Fujita K, Tani J, Masaki T (2021) MicroRNAs in the pathogenesis of hepatocellular carcinoma: a review. Cancers 13(3):514. https://doi.org/10.3390/cancers13030514
Niu X, Wei N, Pen L et al (2022) miR-34a-5p plays an inhibitory role in hepatocellular carcinoma by regulating target gene VEGFA. Malays J Pathol 44(1):39–52
Nomura F, Yaguchi M, Togawa A et al (2000) Enhancement of poly-adenosine diphosphate-ribosylation in human hepatocellular carcinoma. J Gastroenterol Hepatol 15(5):529–535. https://doi.org/10.1046/j.1440-1746.2000.02193.x
Pang SW, Armon S, Chook JB et al (2024) Association of Fusobacterium Nucletum infection with the clinicopathological characteristics in colorectal cancer patients. Mol Biol Rep 51(1):124. https://doi.org/10.1007/s11033-023-09150-5
Parzych KR, Klionsky DJ (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal20(3): 460 – 73. https://doi.org/10.1089/ars.2013.5371
Qiu D, Wang G, Chen L et al (2014) The expression of beclin-1, an autophagic gene, in hepatocellular carcinoma associated with clinical pathological and prognostic significance. BMC Cancer 14:327. https://doi.org/10.1186/1471-2407-14-327
Qiu G, Li X, Che X et al (2015) SIRT1 is a regulator of autophagy: implications in gastric cancer progression and treatment. FEBS Lett 589(16):2034–2042. https://doi.org/10.1016/j.febslet.2015.05.042
Rodríguez-Vargas JM, Rodríguez MI, Majuelos-Melguizo J et al (2016) Autophagy requires poly(adp-ribosyl)ationdependent AMPK nuclear export. Cell Death Differ 23(12):2007–2018. https://doi.org/10.1038/cdd.2016.80
Santana-Codina N, Mancias JD, Kimmelman AC (2017) The role of autophagy in cancer. Annu Rev Cancer Biol 1:19–39. https://doi.org/10.1146/annurev-cancerbio-041816-122338
Schläfli AM, Berezowska S, Adams O et al (2015) Reliable LC3 and p62 autophagy marker detection in formalin fixed paraffin embedded human tissue by immunohistochemistry. Eur J Histochem 59(2):2481. https://doi.org/10.4081/ejh.2015.2481
Schreiber V, Dantzer F, Ame JC et al (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7(7):517–528. https://doi.org/10.1038/nrm1963
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Takamura A, Komatsu M, Hara T et al (2011) Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25(8):795–800. https://doi.org/10.1101/gad.2016211
Tobita T, Guzman-Lepe J, Takeishi K et al (2016) SIRT1 disruption in human fetal hepatocytes leads to increased accumulation of glucose and lipids. PLoS ONE 11(2):e0149344. https://doi.org/10.1371/journal.pone.0149344
Umemura A, He F, Taniguchi K et al (2016) p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell 29(6):935–948. https://doi.org/10.1016/j.ccell.2016.04.006
Wang RH, Sengupta K, Li C (2008) Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14(4):312–323. https://doi.org/10.1016/j.ccr.2008.09.001
Wang Y, Cui R, Zhang X et al (2016) SIRT1 increases YAP- and MKK3-dependent p38 phosphorylation in mouse liver and human hepatocellular carcinoma. Oncotarget 7(10):11284–11298. https://doi.org/10.18632/oncotarget.7022
White E (2015) The role of autophagy in cancer. J Clin Invest 125(1):42–46. https://doi.org/10.1172/JCI73941
Wong MMT, Chan HY, Aziz NA et al (2021) Interplay of autophagy and cancer stem cells in hepatocellular carcinoma. Mol Biol Rep 2021:1–23. https://doi.org/10.1007/s11033-021-06334-9
Wu DH, Jia CC, Chen J et al (2014) Autophagic LC3B overexpression correlates with malignant progression and predicts a poor prognosis in hepatocellular carcinoma. Tumour Biol 35(12):12225–12233. https://doi.org/10.1007/s13277-014-2531-7
Xi SY, Lu JB, Chen JW et al (2013) The stone-like pattern of LC3A expression and its clinicopathologic significance in hepatocellular carcinoma. Biochem Biophys Res Comm 431(4):760–766. https://doi.org/10.1016/j.bbrc.2012.12.151
Yang Z, Klionsky DJ (2009) An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 335:1–32. https://doi.org/10.1007/978-3-642-00302-8_1
Yang XD, Kong FE, Qi L et al (2021) PARP inhibitor Olaparib overcomes Sorafenib resistance through reshaping the pluripotent transcriptome in hepatocellular carcinoma. Mol Cancer 20(1):20. https://doi.org/10.1186/s12943-021-01315-9
Yu LX, Zhang BL, Yang MY et al (2019) MicroRNA-106b-5p promotes hepatocellular carcinoma development via modulating FOG2. OncoTargets Ther 12:5639–5647. https://doi.org/10.2147/OTT.S203382
Acknowledgements
Magdelyn Wong is the recipient of Sunway University Postgraduate’s Degree by Research Studentship. We also thank Wenzhou-Kean University for continuously supporting the research work.
Funding
This work was partly funded by Ministry of Higher Education Malaysia (FRGS/1/2019/SKK08/SYUC/02/1) and Sunway Medical Centre Research Funds (SRC/002/2017/FR and SRC/003/ 2017/ FR). We also thank Wenzhou-Kean University (WKU) Student Partnering with Faculty (SpF) Research Program [grant number WKUSPF2023025 and WKUSPF2023026], Internal Start-Up Research Grant [grant number ISRG2023030] and Zhejiang Province Department of Education General Program 2023 [grant number Y202353358] for partly supporting this work.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design as well as the analysis and interpretation of the data. Material preparation and data collection were performed by Magdelyn Mei-Theng Wong. The first draft of the manuscript was written by Magdelyn Mei-Theng Wong and Sin-Yeang Teow. Sin-Yeang Teow reviewed and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Ethical approval for this study was granted from Sunway University Research Ethics Committee (SUNREC 2017/051) and Sunway Medical Centre Independent Research Ethics Committee (013/2017/ER). Archived FFPE tissue blocks of HCC patients from year 2012 to 2016 from Sunway Medical Centre were used in this project. The written informed consent was waived with the approval of the Institutional Review Board (IRB).
Conflict of interest
The authors declare no conflict of interest.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wong, M.MT., Aziz, N.A., Ch’ng, E.S. et al. Expression of LC3A, LC3B and p62/SQSTM1 autophagy proteins in hepatocellular carcinoma (HCC) tissues and the predicted microRNAs involved in the autophagy-related pathway. J Mol Histol (2024). https://doi.org/10.1007/s10735-024-10191-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10735-024-10191-8