Abstract
Background
The prevalence of TNBC in India is higher compared to western countries. There is a multitude of biomarkers associated with different clinical outcomes of TNBC with contradictory reports. Identification of a set of specific biomarkers from the very many number of proteins reported in the literature to predict prognosis of TNBC is an urgent clinical need.
Methodology
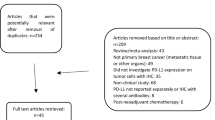
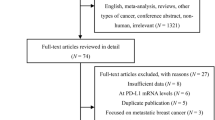
A systematic review of key molecular biomarkers in cohort studies that have been investigated for their role in breast cancer prognosis was conducted. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) methodology was followed. A meta-analysis was used to evaluate their pooled hazard ratio (HR) and the corresponding 95% confidence interval (95% CI) statistically. Immunohistochemical characterization of the meta-analyzed markers were performed in a cohort of 200 retrospective TNBC and 100 non TNBC patient tissues. Kaplan–Meier plot were used to evaluate disease free survival (DFS), and overall survival (OS). Cox regression models were used to evaluate predictors of DFS and OS.
Results
Using a meta-analytical approach, we consolidated the biomarker signatures associated with survival outcomes in breast cancers. The promising markers that emerged for the prediction of DFS and OS included E-Cadherin, Survivin, p53, MTA1, HIF1A, CD133, Vimentin and CK5/6. Evaluation of these markers in tumor tissue revealed that subcellular localization of p53, MTA1 and HIF1A had a significant association in predicting TNBC prognosis. Kaplan Meier plot revealed that p53 (OS p = 0.007, DFS p = 0.004), HIF 1 A (OS p = 0.054, DFS p = 0.009) and MTA1 (OS p = 0.043, DFS = p = 0.001) expression in the primary tumor tissue were found to be significantly correlated with poor OS and DFS, whereas expression of Survivin (DFS p = 0.024) and E Cadherin (DFS p = 0.027) correlated with DFS alone in TNBC. Univariate analysis revealed that p53, HIF1A and MTA1 could be independent prognostic markers.
Conclusion
Our study suggests cytoplasmic over expression of HIF1A, nuclear over expression of MTA1 and mutated p53 in the primary tumor tissue of TNBC have significance as markers predicting survival of TNBC patients.






Similar content being viewed by others
References
Assidicky R, Tokat UM, Tarman IO, Saatci O, Ersan PG, Raza U et al (2022) Targeting HIF1-alpha/miR-326/ITGA5 axis potentiates chemotherapy response in triple-negative breast cancer. J Breast cancer Res 193(2):331–348
Cheng C-W, Liu Y-F, Yu J-C, Wang H-W, Ding S-L, Hsiung C-N et al (2012) Prognostic significance of cyclin D1, β-catenin, and MTA1 in patients with invasive ductal carcinoma of the breast. J Annals Surg Oncol 19:4129–4139
da Silva JL, Rodrigues FR, de Mesquita GG, Fernandes PV, Thuler LCS, de Melo AC et al (2021) Triple-negative breast cancer: assessing the role of immunohistochemical biomarkers on neoadjuvant treatment. J Breast Cancer: Targets 31–44
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA et al (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. J Clin cancer Res 13(15):4429–4434
Geyer FC, Pareja F, Weigelt B, Rakha E, Ellis IO, Schnitt SJ et al (2017) The spectrum of triple-negative breast disease: high-and low-grade lesions. J Am J Pathol 187(10):2139–2151
Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A et al (2020) Visualizing and interpreting cancer genomics data via the Xena platform. 38(6):675–678
Goto Y, Thike AA, Ong CCH, Lim JX, Nasir NDM, Li H et al (2020) Characteristics, behaviour and role of biomarkers in metastatic triple-negative breast cancer. J J Clin Pathol 73(3):147–153
Hannafon BN, Gin AL, Xu Y-F, Bruns M, Calloway CL, Ding W-Q (2019) Metastasis-associated protein 1 (MTA1) is transferred by exosomes and contributes to the regulation of hypoxia and estrogen signaling in breast cancer cells. J Cell Communication Signal 17(1):1–13
Jang KS, Paik SS, Chung H, Oh YH, Kong G (2006) MTA1 overexpression correlates significantly with tumor grade and angiogenesis in human breast cancers. J Cancer Sci 97(5):374–379
Kulkarni A, Kelkar DA, Parikh N, Shashidhara LS, Koppiker CB, Kulkarni M (2020) Meta-analysis of prevalence of triple-negative breast cancer and its clinical features at incidence in Indian patients with breast cancer. J JCO Global Oncol 6:1052–1062
Kumar S, Bal A, Das A, Bhattacharyya S, Laroiya I, Khare S et al (2021) Molecular subtyping of triple negative breast cancer by surrogate immunohistochemistry markers. J Appl Immunohistochem Mol Morphology 29(4):251–257
Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA et al (2008) Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J J Clin Oncol 26(8):1275–1281
Mitri ZI, Abuhadra N, Goodyear SM, Hobbs EA, Kaempf A, Thompson AM et al (2022) Impact of TP53 mutations in Triple negative breast Cancer. J NPJ Precision Oncol 6(1):64
Skinner KE, Haiderali A, Huang M, Schwartzberg LS (2021) Real-world effectiveness outcomes in patients diagnosed with metastatic triple-negative breast cancer. J Future Oncol 17(8):931–941
Sporikova Z, Koudelakova V, Trojanec R, Hajduch M (2018) Genetic markers in triple-negative breast cancer. J Clin Breast cancer 18(5):e841–e50
Srivastava A (2017) Mammographic screening or breast cancer awareness? Time to ponder. J Indian J Surg 79(5):446–449
Stewart RL, Updike KL, Factor RE, Henry NL, Boucher KM, Bernard PS et al (2019) A multigene assay determines risk of recurrence in patients with triple-negative breast cancer. J Cancer Res 79(13):3466–3478
Thakur KK, Bordoloi D, Kunnumakkara AB (2018) Alarming burden of triple-negative breast cancer in India. J Clin Breast cancer 18(3):e393–e9
Thike AA, Iqbal J, Cheok PY, Chong APY, Tse GM-K, Tan B et al (2010) Triple negative breast cancer: outcome correlation with immunohistochemical detection of basal markers. J Am J Surg Pathol 34(7):956–964
Tseng L, Hsu N, Chen S, Lu Y, Lin C, Chang D et al (2013) Distant metastasis in triple-negative breast cancer. J Neoplasma 60(3):290–294
Turner NC, Telli ML, Rugo HS, Mailliez A, Ettl J, Grischke E-M et al (2019) A phase II study of talazoparib after platinum or cytotoxic nonplatinum regimens in patients with advanced breast cancer and germline BRCA1/2 mutations (ABRAZO). J Clin Cancer Res 25(9):2717–2724
Untch M, Harbeck N, Huober J, von Minckwitz G, Gerber B, Kreipe H-H et al (2015) Primary therapy of patients with early breast cancer: evidence, controversies, consensus. 75(06):556–565
Wu M, Wen L, Zhou Y, Wu W (2022) Role of lncRNA AGAP2-AS1 in breast cancer cell resistance to apoptosis by the regulation of MTA1 promoter activity. J Technol cancer Res Treat 21:15330338221085361
Zhang S-B, Tang Q-R (2016) Predicting protein subcellular localization based on information content of gene ontology terms. J Comput Biology Chem 65:1–7
Acknowledgements
The authors wish to acknowledge CSIR for providing fellowship to SSS.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and publication of this article: This research was funded by DBT, grant number BT/PR18812/COE/34/01/2017, dated 19-05-2017.
Author information
Authors and Affiliations
Contributions
KS supervised the experiments and reviewed the manuscript. SSS performed experiments and drafted the manuscript. JK supported statistics and provided essential knowledge on statistical methods. AL edited the manuscript. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All experiments were approved by the committee for Research and Ethics of the local authorities The Institutional Review Board (IRB No. 06/2017/02) and the Human ethical committee of the Regional Cancer Centre (HEC 20/2017) have approved the study.
Consent to publish
Not applicable.
Competing Interests
The authors declared no potential conflicts of interest concerning this article’s research, authorship, and publication.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sharaf, S.S., Jaganath Krishna, K.M., Lekshmi, A. et al. Subcellular expression of MTA1, HIF1A and p53 in primary tumor predicts aggressive triple negative breast cancers: a meta-analysis based study. J Mol Histol (2024). https://doi.org/10.1007/s10735-024-10190-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10735-024-10190-9