Abstract
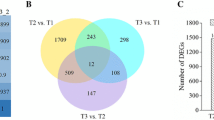
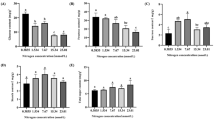
Nitrogen (N) is an important macronutrient for blueberry growth and development. To date, the response mechanism of blueberry to N deficiencystress has not been well elaborated. In this study, the adaptability strategy of blueberries under nitrogen deficiency stress was comprehensively studied using one-year-old seedlings of the highbush blueberry cultivar 'Legacy' subjected to two treatments: normal N fertilization treatment as a control (CK, 15 mM N fertilization) and N deficiency treatment (ND, without N fertilization). The physiological results showed that under nitrogen deficiency stress, plant growth slowed, the dry and fresh weights of the plants decreased, and the chlorophyll (Chl) content decreased significantly. The contents of hydrogen peroxide (H2O2), malonaldehyde (MDA), reduced glutathione (GSH), and the activity of superoxide dismutase (SOD) were increased. The absorption of nutrients was significantly affected under nitrogen deficiency treatment. The N, phosphorus (P), potassium (K), sodium (Na) and iron (Fe) contents were decreased, while calcium (Ca), magnesium (Mg), copper (Cu) and zinc (Zn) contents were increased. A total of 1465 differentially expressed genes (DEGs) were obtained in transcriptomic analysis, of which N metabolism-, carbohydrate metabolism- and amino acid metabolism-related pathways were significantly enriched and involved in efficient N uptake and transport. Glycolysis/gluconeogenesis (EMP) and the pentose phosphate pathway (PPP) are the main energy sources in the process of carbohydrate metabolism, and the five upregulated DEGs may play key regulatory roles. We also identified DEGs involved in ions transport, phenylpropanoid biosynthesis pathway, flavonoid biosynthesis pathway, hormone signaling and related transcription factors (TFs). Our results reveal the regulatory mechanisms of the blueberry response to nitrogen deficiency stress and provide new strategies for N nutrient management in blueberry cultivation.






Similar content being viewed by others
Abbreviations
- Chl:
-
Chlorophyll
- H2O2 :
-
Hydrogen peroxide
- MDA:
-
Malondialdehyde
- AsA:
-
Ascorbic acid
- GSH:
-
Reduced glutathione
- SOD:
-
Superoxide dismutase
- POD:
-
Peroxidase
- CAT:
-
Catalase
- C:
-
Carbon
- N:
-
Nitrogen
- DEGs:
-
Differentially expressed genes
- EMP:
-
Glycolysis/gluconeogenesis
- PPP:
-
The pentose phosphate pathway
- PGM:
-
Phosphoglucomutase
- PFK5:
-
ATP-dependent 6-phosphofructokinase
- FBA5:
-
Fructose-bisphosphate aldolase
- gpmA:
-
2,3-Bisphosphoglycerate-dependent phosphoglycerate mutase
- ENO1:
-
Enolase 1
- OsI:
-
Pyruvate kinase
- ADH:
-
Alcohol dehydrogenase
- G6PD2:
-
Glucose-6-phosphate 1-dehydrogenase
- At2g20420:
-
Succinate-CoA ligase
- NR:
-
Nitrate reductase
- NiR:
-
Nitrite reductase
- NRTs:
-
Nitrate transporters
- GS:
-
Glutamine synthetase
- GOGAT:
-
Glutamate synthase
- TFs:
-
Transcription factors
- WRKY:
-
WRKYGQK
- MYB:
-
Myeloblastosis
- bHLH:
-
Basic helix-loop-helix
- ERF:
-
Ethylene response factor
- ZAT:
-
Cys2/His2 zinc finger
- NAC:
-
NAM, ATAF1/2, and CUC2
- RAV:
-
Related to ABI3/VP1
- NLP:
-
NIN-like protein
- AP2:
-
Apetala2
- DOF:
-
DNA binding with one finger
- P:
-
Phosphorus
- K:
-
Potassium
- Ca:
-
Calcium
- Mg:
-
Magnesium
- Fe:
-
Iron
- Mn:
-
Manganese
- Cu:
-
Copper
- Zn:
-
Zinc
- CNGC:
-
Cyclic nucleotide-gated channel
- CBL/CIPK:
-
Calcineurin B-like protein and CBL-interacting protein kinase
- CDPK:
-
Calcium-dependent protein kinase
- CaM:
-
Calmodulin
- CML:
-
CaM-like protein
- HAK/KUP/KT:
-
High-affinity K transporter/K uptake permease/K transporter
- HKT:
-
High-affinity K transporter
- CPA:
-
Cation proton antiporter
- KCO/TPK:
-
K channel, Ca2+-activated, outward rectifying and two-pore K channel
- PHT:
-
Phosphate transporter
- ZIP:
-
Zinc-regulated transporter
- YSL:
-
Yellow stripe-like
- HMA:
-
Heavy metal ATPase
- CDF:
-
Cation diffusion facilitator
- NRAMP:
-
Natural resistance associated macrophage protein
- IRT:
-
Iron-regulated transporter
- FRO:
-
Ferric chelate reductase
- COPT:
-
Copper transporter
- CCH/ATX/COX/CCS:
-
Copper chaperone/antioxidant protein/copper chaperone for cytochrome c oxidase/copper chaperone for Cu/Zn superoxide dismutase
- MGT:
-
Magnesium transporter
References
An JP, Zhang XW, Xu RR, You CX, Wang XF, Hao YJ (2018) Apple MdERF4 negatively regulates salt tolerance by inhibiting MdERF3 transcription. Plant Sci 276:181–188. https://doi.org/10.1016/j.plantsci.2018.08.017
Anderson ME (1985) Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol 113:548–555. https://doi.org/10.1016/s0076-6879(85)13073-9
Aranjuelo I, Tcherkez G, Molero G, Gilard F, Avice JC, Nogués S (2013) Concerted changes in N and C primary metabolism in alfalfa (Medicago sativa) under water restriction. J Exp Bot 64:885–897. https://doi.org/10.1093/jxb/ers367
Bouguyon E, Perrine-Walker F, Pervent M, Rochette J, Cuesta C, Benkova E, Martinière A, Bach L, Krouk G, Gojon A, Nacry P (2016) Nitrate controls root development through posttranscriptional regulation of the NRT1.1/NPF6.3 transporter/sensor. Plant Physiol 172:1237–1248. https://doi.org/10.1104/pp.16.01047
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1006/abio.1976.9999
Bryla DR, Strik BC (2015) Nutrient requirements, leaf tissue standards, and new options for fertigation of northern highbush blueberry. HortTechnol 25:464–470. https://doi.org/10.21273/HORTTECH.25.4.464
Buchanan-Wollaston V (1997) The molecular biology of leaf senescence. J Exp Bot 48:181–199. https://doi.org/10.1093/jxb/48.2.181
Chen Q, Liu Z, Wang B, Wang X, Lai J, Tian F (2015) Transcriptome sequencing reveals the roles of transcription factors in modulating genotype by nitrogen interaction in maize. Plant Cell Rep 34:1761–1771. https://doi.org/10.1007/s00299-015-1822-9
Chen SF, Zhou YQ, Chen Y, Gu J (2018a) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:884–890. https://doi.org/10.1093/bioinformatics/bty560
Chen XG, Wang J, Wang ZL, Li WY, Wang CY, Yan SH, Li HM, Zhang AJ, Tang ZH, Wei M (2018b) Optimized nitrogen fertilizer application mode increased culms lignin accumulation and lodging resistance in culms of winter wheat. Field Crop Res 228:31–38. https://doi.org/10.1016/j.fcr.2018.08.019
Conesa BA, Götz S, García-Gómez J, Terol J, Talón M, Robles (2005) Blast2GO: a universal tool for annotation visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Coruzzi GM, Zhou L (2001) Carbon and nitrogen sensing and signaling in plants: emerging “matrix effects.” Curr Opin Plant Biol 4:247–253. https://doi.org/10.1016/s1369-5266(00)00168-0
Coskun D, Britto DT, Shi WM, Kronzucker HJ (2017) Nitrogen transformations in modern agriculture and the role of biological nitriication inhibition. Native Plants 3:17074. https://doi.org/10.1038/nplants.2017.74
Dai XY, Wang YY, Zhang WH (2016) OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J Exp Bot 67:947–960. https://doi.org/10.1093/jxb/erv515
Dai J, Wang NQ, Xiong HC, Qiu W, Nakanishi KT, Nishizawa NK, Zuo YM (2018) The yellow stripe-like (YSL) gene functions in internal copper transport in peanut. Genes 9:635. https://doi.org/10.3390/genes9120635
Darnell RL, Casamali B, Williamson JG (2015) Nutrient assimilation in southern highbush blueberry and implications for the field. HortTechnol 25:460–463. https://doi.org/10.21273/HORTTECH.25.4.460
Fothergill-Gilmore LA (1986) The evolution of the glycolytic pathway. Trends Biochem Sci 11:47–51. https://doi.org/10.1016/0968-0004(86)90233-1
Gao SJ, Wang SY, Hu YN, Chen SY, Guo JX, Shi LX (2022) Comparative differences in photosynthetic characteristics, ion balance, and nitrogen metabolism between young and old wild soybean leaves under nitrogen deficiency. Plant Stress 4:100064. https://doi.org/10.1016/j.stress.2022.100064
Ge M, Wang YC, Liu YH, Jiang L, He B, Ning LH, Du HY, Lyu YD, Zhou L, Lin F, Zhang TF, Liang SQ, Lu HY, Zhao H (2020) The NIN-like protein 5 (ZmNLP5) transcription factor is involved in modulating the nitrogen response in maize. Plant J 102:353–368. https://doi.org/10.1111/tpj.14628
González-Vega RI, Cárdenas-López JL, López-Elías JA, Ruiz-Cruz S, Reyes-Díaz A, Perez-Perez LM, Cinco-Moroyoqui FJ, Robles-Zepeda RE, Borboa-Flores J, Del-Toro-Sánchez CL (2021) Optimization of growing conditions for pigments production from microalga Navicula incerta using response surface methodology and its antioxidant capacity. Saudi J Biol Sci 28:1401–1416. https://doi.org/10.1016/j.sjbs.2020.11.076
Gu JJ, Xu JH, Guo YP, Zong Y, Chen WR, Guo WD (2021) Cloning and functional analysis of ZIP transporters in blueberry. Sci Hortic 278:109871. https://doi.org/10.1016/j.scienta.2020.109871
Guo HY, Wang YC, Wang LQ, Hu P, Wang YM, Jia YY, Zhang CR, Zhang Y, Zhang YM, Wang C, Yang CP (2017) Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol J 15:107–121. https://doi.org/10.1111/pbi.12595
Hageman RH, Flesher D (1960) Nitrate reductase activity in corn seedlings as affected by light and nitrate content of nutrient media. Plant Physiol 35:700–708. https://doi.org/10.1104/pp.35.5.700
Hasanuzzaman M, Bhuyan MHMB, Zulfifiqar F, Raza A, Mohsin SM, Mahmud JA, Fujita M, Fotopoulos V (2020) Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants 9:681. https://doi.org/10.3390/antiox9080681
Hodges DM, DeLong JM, Forney CF, Prange RK (1999) Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207:604–611. https://doi.org/10.1007/s004250050524
Hussain D, Haydon MJ, Wang Y, Edwin Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS (2004) P-Type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16:1327–1339. https://doi.org/10.1105/tpc.020487
Kavanová M, Gloser V (2005) The use of internal nitrogen stores in the rhizomatous grass Calamagrostis epigejos during regrowth after defoliation. Ann Bot-London 95:457–463. https://doi.org/10.1093/aob/mic054
Khan MN, Mobin M, Abbas ZK, Siddiqui MH (2017) Nitric oxide-induced synthesis of hydrogen sulfide alleviates osmotic stress in wheat seedlings through sustaining antioxidant enzymes, osmolyte accumulation and cysteine homeostasis. Nitric Oxide 68:91–102. https://doi.org/10.1016/j.niox.2017.01.001
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. https://doi.org/10.1038/nmeth.3317
Korcak RF, Galletta GJ, Draper DA (1982) Response of blueberry seedlings to a range of soil types. J Amer Soc Hort Sci 107:1153–1160
Law MY, Charles SA, Halliwell B (1983) Glutathione and ascorbic acid in spinach (Spinacia oleracea) chloroplasts. The effect of hydrogen peroxide and of Paraquat. Biochem J 210:899–903. https://doi.org/10.1042/bj2100899
Lea PJ, Miflin BJ (1974) Alternative route for nitrogen assimilation in higher plants. Nature 251:614–616. https://doi.org/10.1038/251614a0
Lea US, Slimestad R, Smedvig P, Lillo C (2007) Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225:1245–1253. https://doi.org/10.1007/s00425-006-0414-x
Lee WJ, Truong HA, Trnh CS, Kim JH, Lee S, Hong SW, Lee H (2020) NITROGEN RESPONSE DEFICIENCY 1-mediated CHL1 induction contributes to optimized growth performance during altered nitrate availability in Arabidopsis. Plant J 104:1382–1398. https://doi.org/10.1111/tpj.15007
Li W, Xiang F, Zhong M, Zhou L, Liu H, Li S, Wang X (2017) Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis). Sci Rep 7:1693. https://doi.org/10.1038/s41598-017-01949-0
Li H, Liu H, Wang Y, Teng RM, Liu JY, Lin SJ, Zhuang J (2020a) Cytosolic ascorbate peroxidase 1 modulates ascorbic acid metabolism through cooperating with nitrogen regulatory protein P-II in tea plant under nitrogen deficiency stress. Genomics 112:3497–3503. https://doi.org/10.1016/j.ygeno.2020.06.025
Li SH, Li YM, Gao Y, He XR, Zhang DL, Liu BB, Li QM (2020) Effects of CO2 enrichment on non-structural carbohydrate metabolism in leaves of cucumber seedlings under salt stress. Sci Hortic 265:109275. https://doi.org/10.1016/j.scienta.2020.109275
Li YD, Pei JB, Li C, Sun HY (2021) 2020 Annual report of blueberry industry in China. J Jilin Agric Univ 43:1–8. https://doi.org/10.13327/j.jjlau.2021.1071
Li XJ, Liu X, Gu MY, Pi BY, Sun SL, Zhao TJ, Yu BJ (2022) Overexpression of GsCNGC20-f from Glycine soja confers submergence tolerance to hairy-root composite soybean plants and Arabidopsis seedlings by enhancing anaerobic respiration. Environ Exp Bot 199:104901. https://doi.org/10.1016/j.envexpbot.2022.104901
Liang Y, Lu J, Zhang L, Wu S, Wu Y (2003) Estimation of black tea quality by analysis of chemical composition and colour difference of tea infusions. Food Chem 80:283–290. https://doi.org/10.1016/S0308-8146(02)00415-6
Liu C, Liao WB (2022) Potassium signaling in plant abiotic responses: crosstalk with calcium and reactive oxygen species/reactive nitrogen species. Plant Physiol Bioch 173:110–121. https://doi.org/10.1016/j.plaphy.2022.01.016
Liu W, Li QW, Wang Y, Wu T, Yang YF, Zhang XZ, Han ZH, Xu XF (2017) Ethylene response factor AtERF72 negatively regulates Arabidopsis thaliana response to iron deficiency. Biochem Biophys Res Commun 491:862–868. https://doi.org/10.1016/j.bbrc.2017.04.014
Liu SR, Cui S, Ying FY, Nasar J, Wang Y, Gao Q (2021) Simultaneous improvement of protein concentration and amino acid balance in maize grains by coordination application of nitrogen and sulfur. J Cereal Sci 99:103189. https://doi.org/10.1016/j.jcs.2021.103189
Majumdar A, Kar RK (2019) Orchestration of Cu-Zn SOD and class III peroxidase with upstream interplay between NADPH oxidase and PM H+-ATPase mediates root growth in Vigna radiata (L.) Wilczek. J Plant Physiol 232:248–256. https://doi.org/10.1016/j.jplph.2018.11.001
Mapodzeke JM, Adil MF, Sehar S, Karim MF, Saddique MAB, Ouyang Y, Shamsi IM (2021) Myriad of physio-genetic factors determining the fate of plant under zinc nutrient management. Environ Exp Bot 189:104559. https://doi.org/10.1016/j.envexpbot.2021.104559
Martin RE, Asner GP, Sack L (2007) Genetic variation in leaf pigment, optical and photosynthetic function among diverse phenotypes of metrosideros polymorpha grown in a common garden. Oecologia 151:387–400. https://doi.org/10.1007/s00442-006-0604-z
Martyn AJ, Thomasa CR, O’Neill ME, Offord CA, Mcconchie R (2007) Bract browning in waratahs (Telopea spp.) is not a localized calcium deficiency disorder. Sci Hortic 112:434–438. https://doi.org/10.1016/j.scienta.2007.03.005
Nemie-Feyissa D, Olafsdottir SM, Heidari B, Lillo C (2014) Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in Arabidopsis leaves. Phytochemistry 98:34–40. https://doi.org/10.1016/j.phytochem.2013.12.006
Norberto S, Silva S, Meireles M, Faria A, Pintado M, Calhau C (2013) Blueberry anthocyanins in health promotion: a metabolic overview. J Funct Foods 5:1518–1528. https://doi.org/10.1016/j.jff.2013.08.015
Ochmian I, Kozos K, Jaroszewska A, Malinowski R (2021) Chemical and enzymatic changes of different soils during their acidification to adapt them to the cultivation of highbush blueberry. Agronomy 11:44. https://doi.org/10.3390/agronomy1101004
Rai S, Singh PK, Mankotia S, Swain J, Satbhai SB (2021) Iron homeostasis in plants and its crosstalk with copper, zinc, and manganese. Plant Stress 1:100008. https://doi.org/10.1016/j.stress.2021.100008
Rout GR, Sahoo S (2015) Role of iron in plant growth and metabolism. Rev Agric Sci 3:1–24. https://doi.org/10.7831/ras.3.1
Shi JW, Yan X, Sun TT, Shen YX, Shi Q, Wang W, Bao MZ, Luo H, Nian FZ, Ning GG (2022) Homeostatic regulation of flavonoid and lignin biosynthesis in phenylpropanoid pathway of transgenic tobacco. Gene 809:146017. https://doi.org/10.1016/j.gene.2021.146017
Sperotto RA, Boff T, Duarte GL, Santos LS, Grusak MA, Fett JP (2010) Identification of putative target genes to manipulate Fe and Zn concentrations in rice grains. J Plant Physiol 167:1500–1506. https://doi.org/10.1016/j.jplph.2010.05.003
Stewart RR, Bewley JD (1980) Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol 65:245–248. https://doi.org/10.1104/pp.65.2.245
Su N, Ling F, Xing AM, Zhao HH, Zhu YW, Wang Y, Deng XP, Wang CF, Xu XM, Hu ZB, Cui J, Shen ZG, Xi Y (2020) Lignin synthesis mediated by CCoAOMT enzymes is required for the tolerance against excess Cu in Oryza sativa. Environ Exp Bot 175:104059. https://doi.org/10.1016/j.envexpbot.2020.104059
Sun L, Di DW, Li GJ, Li YL, Kronzucker HJ, Shi WM (2020) Transcriptome analysis of rice (Oryza sativa L.) in response to ammonium resupply reveals the involvement of phytohormone signaling and the transcription factor OsJAZ9 in reprogramming of nitrogen uptake and metabolism. J Plant Physiol 246–247:153137. https://doi.org/10.1016/j.jplph.2020.153137
Sun TT, Zhang JK, Zhang Q, Li XL, Li MJ, Yang YZ, Zhou J, Wei QP, Zhou BB (2021) Integrative physiological, transcriptome, and metabolome analysis reveals the effects of nitrogen sufficiency and deficiency conditions in apple leaves and roots. Environ Exp Bot 192:104633. https://doi.org/10.1016/j.envexpbot.2021.104633
Tang RJ, Sheng L (2017) Regulation of calcium and magnesium homeostasis in plants: from transporters to signaling network. Curr Opin Plant Biol 39:97–105. https://doi.org/10.1016/j.pbi.2017.06.009
Tewari RK, Kumar P, Sharma PN (2007) Oxidative stress and antioxidant responses in young leaves of mulberry plants grown under nitrogen, phosphorus or potassium deficiency. J Integr Plant Biol 49:313–322. https://doi.org/10.1111/j.1744-7909.2007.00358.x
Tripathi DK, Singh S, Gaur S, Singh S, Yadav V, Liu S, Singh VP, Sharma S, Srivastava P, PrasadSM DNK, Chauhan DK, Sahi S (2018) Acquisition and homeostasis of iron in higher plants and their probable role in abiotic stress tolerance. Front Env Sci 5:86. https://doi.org/10.3389/fenvs.2017.00086
Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W (2012) Metabolic engineering of novel lignin in biomass crops. New Phytol 196:978–1000. https://doi.org/10.1111/j.1469-8137.2012.04337.x
Wang W, Hu B, Yuan DY, Liu YQ, Che RH, Hu YC, Ou SJ, Liu YX, Zhang ZH, Wang HR, Li H, Jiang ZM, Zhang ZL, Gao XK, Qiu YH, Meng XB, Liu YX, Bai Y, Liang Y, Wang YQ, Zhang LH, Li LG, Sodmergen JHC, Li JY, Chu CC (2018) Expression of the nitrate transporter gene OsNRT1. 1A/OsNPF6. 3 confers high yield and early maturation in rice. Plant Cell 30:638–651. https://doi.org/10.1105/tpc.17.00809
Wang M, Zhang PL, Liu Q, Li GJ, Di DW, Xia GM, Kronzucker HJ, Fang S, Chu JF, Shi WM (2020) TaANR1-TaBG1 and TaWabi5-TaNRT2s/NARs link ABA metabolism and nitrate acquisition in wheat roots. Plant Physiol 182:1440–1453. https://doi.org/10.1104/pp.19.01482
Wu ZC, Jiang Q, Yan T, Zhang X, Xu SJ, Shi HZ, Deng T, Li FR, Du YQ, Du RY, Hu CX, Wang X, Wang FH (2020) Ammonium nutrition mitigates cadmium toxicity in rice (Oryza sativa L.) through improving antioxidase system and the glutathione-ascorbate cycle efficiency. Ecotox Environ Safety 189:11001. https://doi.org/10.1016/j.ecoenv.2019.110010
Wu MX, Zou Y, Yu YH, Chen BX, Zheng QW, Ye ZW, Wei T, Ye SQ, Guo LQ, Lin JF (2021) Comparative transcriptome and proteome provide new insights into the regulatory mechanisms of the postharvest deterioration of Pleurotus tuoliensis fruitbodies during storage. Food Res Int 147:110540. https://doi.org/10.1016/j.foodres.2021.110540
Xu L, Jin L, Long L, Liu LL, He X, Gao W, Zhu LF, Zhang XL (2012) Overexpression of GbWRKY1 positively regulates the Pi starvation response by alteration of auxin sensitivity in Arabidopsis. Plant Cell Rep 31:2177–2188. https://doi.org/10.1007/s00299-012-1328-7
Yang HY, Wu YQ, Wu WL, Lyu LF, Li WL (2021) Transcriptomic analysis of blackberry plant (Rubus spp.) reveals a comprehensive metabolic network involved in fruit ripening process. Biologia 76:3827–3840. https://doi.org/10.1007/s11756-021-00896-6
Yang HY, Wu YQ, Duan YK, Zhang CH, Huang ZJ, Wu WL, Lyu LF, Li WL (2022) Metabolomics combined with physiological and transcriptomic analyses reveal regulatory features associated with blueberry growth in different soilless substrates. Sci Hortic 302: 111145. https://doi.org/10.1016/j.scienta.2022.111145
Zhang TT, Kang H, Fu LL, Sun WJ, Hao YJ (2021) NIN-like protein 7 promotes nitrate-mediated lateral root development by activating transcription of tryptophan aminotransferase related 2. Plant Sci 303:110771. https://doi.org/10.1016/j.plantsci.2020.110771
Zhang Z, Wu ZH, Yuan YY, Zhang JX, Wei J, Wu B (2022) Sulfur dioxide mitigates oxidative damage by modulating hydrogen peroxide homeostasis in postharvest table grapes. Postharvest Biol Tec 188:111877. https://doi.org/10.1016/j.postharvbio.2022.111877
Zifkin M, Jin A, Ozga JA, Zaharia LI, Schernthaner JP, Gesell A, Abrams SR, Kennedy JA, Constabel CP (2012) Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol 158:200–224. https://doi.org/10.1104/pp.111.180950
Funding
This work was supported by Jiangsu Agricultural Science and Technology Innovation Fund Project (CX(21)3172), the earmarked fund for Jiangsu Agricultural Industry Technology System (JATS[2021]511), and The “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS〔2021〕021).
Author information
Authors and Affiliations
Contributions
HY: Investigation, Formal analysis, Software, Validation, Writing—original draft, Writing—review & editing. YD, YW, CZ and LL: Formal analysis and Validation. WL and WW: Resources, Funding acquisition and Writing—review & editing. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Communicated by Fei Dai.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

10725_2023_1038_MOESM1_ESM.jpg
Supplementary file1 (JPG 1822 KB)—Fig. S1 A Gene ontology (GO) classification of DEGs between CK and ND. B Number of DEGs involved in KEGG pathway annotation. C Heatmap of DEGs involved in glycolysis/gluconeogenesis (EMP), pentose phosphate pathway (PPP) and citrate (TCA) cycle

Supplementary file2 (JPG 1621 KB)—Fig. S2 Heatmap of DEGs involved in flavonoid biosynthesis pathway

10725_2023_1038_MOESM3_ESM.jpg
Supplementary file3 (JPG 766 KB)—Fig. S3 Comparison of the expression of 8 nitrogen metabolism-related genes between RNA-Seq (A) and qRT-PCR (B)
10725_2023_1038_MOESM7_ESM.xlsx
Supplementary file7 (XLSX 21 KB)—Table S4 DEGs involved in glycine, serine and threonine metabolism and cysteine and methionine metabolism pathways
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, H., Duan, Y., Wu, Y. et al. Physiological and transcriptional responses of carbohydrate and nitrogen metabolism and ion balance in blueberry plants under nitrogen deficiency. Plant Growth Regul 101, 519–535 (2023). https://doi.org/10.1007/s10725-023-01038-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-023-01038-5