Abstract
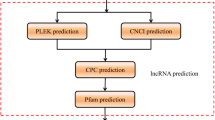
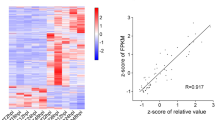
Long non-coding RNAs (lncRNAs) belong to a family of regulatory RNAs of non-coding RNAs (ncRNAs). They play essential roles in plant various developmental processes and stress responses. However, lncRNA functions in cotton are rarely known under attack of cotton aphids (Aphis gossypii Glover). Herein, we performed whole-transcriptome RNA sequencing for cotton four-leaf-stage leaves (Gossypium hirsutum, ZM-03199) damaged by cotton aphids at different time stages (6, 12, 24, 48, and 72 h) and without cotton aphid damage (0 h). We identified 8414 lncRNAs in 6 RNA-Seq data, including 6516 intergenic lncRNAs (77%), 759 anti-sense lncRNAs (7%), 334 intronic lncRNAs (4%) and 805 (10%) sense lncRNAs. Furthermore, 1331 lncRNAs showed differential expressions, and the gene ontology (Go) and KEGG analysis discovered that many target protein-coding genes of lncRNAs were enriched in stress-related categories. In addition, a total of 55 lncRNAs among 8414 lncRNAs were predicted as target mimics of cotton 285 miRNAs. Real-time quantitative PCR (qPCR) confirmed that all selected lncRNAs were relative to aphid damage. These results laid a solid foundation for the study of cotton lncRNA functions under the damage of cotton aphids.








Similar content being viewed by others
References
Akhtar ZR, Tian JC, Chen Y, Fang Q, Hu C, Peng YF, Ye GY (2013) Impact of six transgenic Bacillus thuringiensis rice lines on four nontarget thrips species attacking rice panicles in the paddy field. Environ Entomol 42:173–180
Akhtar ZR, Irshad U, Majid M, Saeed Z, Khan H, Anjum AA, Noreen A, Salman MA, Khalid J, Abubakar M (2018) Risk assessment of transgenic cotton against non-target whiteflies, thrips, jassids and aphids under field conditions in Pakistan. Int J Curr Microbiol App Sci 6:11–24
Blackman RL, Eastop VF (2000) Aphids on the world’s crops, an identification and information guide, 2nd edn. Wiley, Chichester
Chekanova JA (2015) Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol 27:207–216
Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Mortazavi A (2016) A survey of best practices for RNA-Seq data analysis. Genome Biol 17:1–19
Cui J, Luan Y, Jiang N, Bao H, Meng J (2017) Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lnc RNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J 89:577–589
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:155–159
Deng F, Zhang X, Wang W, Yuan R, Shen F (2018) Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress. BMC Plant Biol 18:1–14
Dubey N, Singh K (2018) Role of NBS-LRR proteins in plant defense [M]//molecular aspects of plant-pathogen interaction. Springer, Singapore, pp 115–138
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2013) Pfam: the protein families database. Nucleic Acids Res 41:D222–D230
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037
Herron GA, Wilson LJ (2017) Can resistance management strategies recover insecticide susceptibility in pests? A case study with cotton aphid Aphis gossypii (Aphididae: Hemiptera) in Australian cotton. Austral Entomol 56:1–13
Hu H, Wang M, Ding Y, Zhu S, Zhao G, Tu L, Zhang X (2018) Transcriptomic repertoires depict the initiation of lint and fuzz fibres in cotton (Gossypium hirsutum L.). Plant Biotechnol J 16:1002–1012
Kong L, Zhang Y, Ye Z, Liu X, Zhao S, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35:W345–W349
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinf 9:1–13
Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, Chen W, Liang X, Shang H, Liu W, Shi C, Xiao G, Gou C, Ye W, Xu X, Zhang X, Wei H, Li Z, Zhang G, Wang J, Liu K, Kohel RJ, Percy G, Yu JZ, Zhu Y, Wang J, Yu S (2014a) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46:567–572
Li J, Wu B, Xu J, Liu C (2014b) Genome-wide identification and characterization of long intergenic non-coding RNAs in Ganoderma lucidum. PLoS ONE 9:1–10
Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, Liang X, Huang G, Percy RG, Liu K, Yang W, Chen W, Du X, Shi C, Yuan Y, Ye W, Liu X, Zhang X, Liu W, Wei H, Wei S, Huang G, Zhang X, Zhu S, Zhang H, Sun F, Wang X, Liang J, Wang J, He Q, Huang L, Wang J, Cui J, Song G, Wang K, Xu X, Yu JZ, Zhu Y, Yu S (2015a) Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol 33:524–530
Li P, Song A, Gao C, Jiang J, Chen S, Fang W, Chen F (2015b) The over-expression of a chrysanthemum WRKY transcription factor enhances aphid resistance. Plant Physiol Biochem 95:26–34
Li J, Zhu L, Hull JJ, Liang S, Daniell H, Jin S, Zhang X (2016) Transcriptome analysis reveals a comprehensive insect resistance response mechanism in cotton to infestation by the phloem feeding insect Bemisia tabaci (whitefly). Plant Biotechnol J 14:1956–1975
Li N, Zhou L, Zhang D, Klosterman SJ, Li T, Gui Y, Kong Z, Ma X, Short DPG, Zhang W, Li J, Subbarao KV, Chen J, Dai X (2018) Heterologous expression of the cotton NBS-LRR gene GbaNA1 enhances verticillium wilt resistance in Arabidopsis. Front Plant Sci 9:1–13
Lin B, Zhuo K, Chen S, Hu L, Sun L, Wang X, Zhang LH, Liao JL (2015) A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol 209:1159–1173
Liu X, Hao L, Li D, Zhu L, Hu S (2015) Long non-coding RNAs and their biological roles in plants. Genom Proteom Bioinf 13:137–147
Lu X, Chen X, Mu M, Wang J, Wang X, Wang D, Yin Z, Fan W, Wang S, Guo L, Ye W (2016) Genome-wide analysis of long noncoding RNAs and their responses to drought stress in cotton (Gossypium hirsutum L.). PLoS ONE 11:1–18
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793
Nejat N, Mantri N (2018) Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses. Crit Rev Biotechnol 38:93–105
Page JT, Huynh MD, Liechty ZS, Grupp K, Stelly D, Hulse AM, Ashrafi H, Deynze AV, Wendel JF, Udall JA (2013) Insights into the evolution of cotton diploids and polyploids from whole-genome re-sequencing. G3-Genes Genom Genet 8:1–35
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo M, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee T, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, Rahman M, Rainville LN, Rambani A, Reddy UK, Rong J, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Deynze AV, Vaslin MFS, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang TZ, Dennis ES, Mayer KFX, Peterson DG, Rokhsar DS, Wang X, Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427
Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, Schier AF (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22:577–591
Pfaffl MW (2001) A new mathematicical model for relative quantification in realtime RT-PCR. Nucleic Acids Res 29:2002–2007
Qin T, Zhao H, Cui P, Albesher N, Xiong L (2017) A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol 175:1–38
Quan MY, Tian JX, Yang XH, Du QZ, Song DY, Wang QS, Zhang CD (2016) Association studies reveal the effect of genetic variation in lncRNA UGTRL and its putative target PtoUGT88A1 on wood formation in Populus tomentosa. Tree Genet Genomes 12:1–12
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowsk B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Storey JD (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat 31:2013–2035
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y (2013) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41:e166
Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, Fierens-Onstenk J, de Both M, Peleman J, Liharska T, Hontelez J, Zabeau M (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol 16:1365–1369
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57
Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Zhen Y, Lin C, Shang H, Zhu S, Zou C, Li Q, Yuan Y, Lu C, Wei H, Gou C, Zheng Z, Yin Y, Zhang X, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu Y, Wang J, Yu S (2012) The draft genome of a diploid cotton Gossypium Raimondii. Nat Genet 44:1098–1103
Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W (2013) CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41:e74
Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW (2014) Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci 111:10359–10364
Wang M, Yuan D, Tu L, Gao W, He Y, Hu H, Wang P, Liu N, Lindsey K, Zhang X (2015) Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.). New Phytol 207:1181–1197
Wang M, Wu HJ, Fang J, Chu C, Wang XJ (2017a) A long noncoding RNA involved in rice reproductive development by negatively regulating osa-miR160. Sci Bull 62:470–475
Wang T, Zhao M, Zhang X, Liu M, Yang C, Chen Y, Zhang WH (2017b) Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J Exp Bot 68:5937–5948
Wang Y, Sheng L, Zhang H, Du X, An C, Xia X, Chen S (2017c) CmMYB19 over-expression improves aphid tolerance in chrysanthemum by promoting lignin synthesis. Int J Mol Sci 18:1–13
Wu J, Baldwin IT (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44:1–24
Wu H, Wang Z, Wang M, Wang X (2013) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 161:1875–1884
Yuan D, Tang Z, Wang M, Gao W, Tu L, Jin X, Chen L, He Y, Zhang L, Zhu L, Li Y, Liang Q, Lin Z, Yang X, Liu N, Jin S, Lei Y, Ding Y, Li G, Ruan X, Ruan Y, Zhang X (2015) The genome sequence of Sea-Island cotton (Gossypium Barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep 5:1–16
Zhang YC, Chen YQ (2013) Long noncoding RNAs: new regulators in plant development. Biochem Biophys Res Commun 436:111–114
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Kirkbride SQ, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ (2015) Sequencing of allotetraploid cotton (Gossypium Hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33:531–537
Zhang L, Wang M, Li N, Wang H, Qiu P, Pei L, Xu Z, Wang T, Gao E, Liu J, Liu S, Hu Q, Miao Y, Lindsey K, Tu L, Zhu L, Zhang X (2017) Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol J 16:1172–1185
Zou CS, Wang Q, Lu C, Yang W, Zhang Y, Cheng H, Feng X, Prosper MA, Song G (2016) Transcriptome analysis reveals long noncoding RNAs involved in fiber development in cotton (Gossypium arboreum). Sci China Life Sci 59:164–171
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31471783).
Author information
Authors and Affiliations
Contributions
ZJM and YYZ designed the research. YZ, MQQ, FP, ZX, SQ carried out the experiments, ZJM and WXP analyzed the experimental data and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, J., Yang, Z., Feng, P. et al. Identification and the potential roles of long non-coding RNAs in cotton leaves damaged by Aphis gossypii. Plant Growth Regul 88, 215–225 (2019). https://doi.org/10.1007/s10725-019-00500-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-019-00500-7