Abstract
Calcium-dependent protein kinases (CDPKs or CPKs), unique to plants and some protists, are involved in growth and developmental processes as well as in defence against diverse environmental stresses. CDPKs are encoded by multi-gene families. Despite extensive studies of the CDPKs in many species, information about the evolutionary history and expression patterns of the CDPK family in the staple crop potato (Solanum tuberosum) remains poorly known. In this study, we performed bioinformatics analysis of the potato whole genome sequence and identified 23 potential CDPK genes. These genes are located in 11, of 12, potato chromosomes. Based on the phylogenetic tree and gene structures, the CDPKs were divided into four subfamilies. To determine their expression, reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis was carried out for the CDPK genes in different organs of potato such as young and mature leaves, stems, young shoots, roots, stolons, swollen stolons, flowers and tubers. The CDPKs were expressed in all the organs analysed, but their expression patterns varied greatly. The expression of some CDPKs was strongly organ specific, for example StCPK13 and StCPK18 was found only/mostly in flowers. In Solanum genotypes differing in resistance to Phytophthora infestans, the expression and activity of CDPKs increased in response to a P. infestans elicitor with different kinetics and intensity. The expression levels and activity of the CDPKs correlated positively with the level of the resistance. Our results support earlier suggestion that CDPKs are involved in potato organ development and defence against stresses. We provide new information about the CDPK gene family in the potato and a perspective on its evolutionary history and biological roles of the individual kinases.
Similar content being viewed by others
Introduction
Calcium plays an important role as a universal second messenger in the control of developmental processes and in signal transduction. Changes in calcium ion concentration are sensed, among others, by calcium-dependent protein kinases (CDPKs, also named CPKs). CDPKs are Ser/Thr protein kinases typically comprising five domains: an N-terminal variable domain, a catalytic domain (CD), an autoinhibitory junction domain (JD), a calmodulin-like domain (CLD), and a C-terminal domain (Hrabak et al. 2003). The N- and C-terminal domains are variable, differing in length and amino-acid composition. Moreover, the N-terminal domain often bears myristoylation sites associated with the subcellular localization of the kinase. The specific functions of CDPKs are determined by these variable domains (Hrabak et al. 2003). In a typical CDPK the calmodulin-like domain harbours four EF-hand motifs for Ca2+ binding, and the binding of Ca2+ activates the kinase (Cheng et al. 2002). CDPKs are important sensors and effectors of the Ca2+ flux in plants. CDPKs have different subcellular locations including cytosol, nucleus, plasma membrane, endoplasmic reticulum, peroxisomes, mitochondrial outer membrane, and oil bodies, indicating their possible diverse functions (Lu and Hrabak 2002).
The CDPKs are multifunctional, which is reflected by the increasing number of their substrates being discovered. Numerous studies have demonstrated that CDPKs play key roles in the regulation of plant growth, development, and abiotic and biotic stress resistance (for review see Klimecka and Muszyńska 2007). Some CDPK genes have been shown to be involved in pollen tube growth (Estruch et al. 1994), root development (Ivashuta et al. 2005) and cell division and differentiation (Yoon et al. 1999).
Individual CDPKs in various species have been assigned defined roles. Thus, tomato LeCDPK2 regulates ethylene biosynthesis in response to wound signalling (Kamiyoshihara et al. 2010); Arabidopsis AtCPK1/2/4/5/11 phosphorylate and thereby activate NADPH oxidase to promote reactive oxygen species (ROS) production in response to abiotic and biotic stimuli (Gao et al. 2013); rice OsCPK4 participates in rice tolerance to salt and drought stress by protecting cellular membranes from lipid peroxidation (Campo et al. 2014), grapevine gene VaCPK29 confers tolerance to heat and osmotic stresses (Dubrovina et al. 2017), whereas ZmCPK11 is involved in touch- and wound-induced pathways in maize (Szczegielniak et al. 2012). CDPKs are widely involved in various types of disease resistance (Boudsocq et al. 2010; Boudsocq and Sheen 2013; Romeis and Herde 2014; Wang et al. 2015). They regulate the plants’ defence response locally and also systemically (Romeis and Herde 2014). AtCDPK1/2 modulate the initiation of programmed cell death, while AtCDPK4/5/6/11 phosphorylate specific WRKY transcription factors to regulate immune gene expression. AtCPK1 plays a positive role in resistance to various pathogens by promoting the salicylic acid (SA) signalling pathway (Coca and San Segundo 2010). NtCDPK2 is involved in the induction of Cf-4/Avr4- and Cf-9/Avr9-dependent hypersensitive response (HR) (Romeis et al. 2001).
The CDPKs have been identified in plants (Harmon et al. 2001; Asano et al. 2005), protists (Billker et al. 2009), oomycetes (Broad Institute of Harvard and MIT 2010), and green algae (McCurdy and Harmon 1992; Baillie et al. 2000), but not in any animal or true fungal genome (Zhang and Choi 2001; Hrabak et al. 2003). Plant genomes carry large numbers of CDPK genes; 34 in Arabidopsis thaliana (Cheng et al. 2002; Hrabak et al. 2003), 41 in diploid cotton (Gossypium raimondii) (Liu et al. 2014), 29 or 31 in rice (Oryza sativa) (Asano et al. 2005; Ray et al. 2007, respectively), 20 in wheat (Triticum aestivum) (Li et al. 2008), 35 or 40 in maize (Zea mays) (Ma et al. 2013; Kong et al. 2013, respectively), 29 in tomato (Solanum lycopersicum) (Wang et al. 2015; Hu et al. 2016), and 30 CDPK genes in poplar (Populus trichocarpa) (Zuo et al. 2013). Slightly less numerous are CDPK gene families in cucumber (Cucumis sativus) and grape (Vitis vinifera) with 19 members in both plants (Xu et al. 2015; Zhang et al. 2015).
Despite extensive studies of CDPKs in many species, still little is known about this gene family in the important crop - potato (Solanum tuberosum). Only five isoforms have been characterized so far. It is known that CDPKs are involved in tuber development (Raíces et al. 2001, 2003). StCDPK1 transcript was observed in mesh bulbs and that of StCDPK2 mainly in the leaves. StCDPK1 also plays a role in gibberellic acid (GA) signaling and influences tuberization (Gargantini et al. 2009) while StCDPK2 is associated with plant growth and development (Giammaria et al. 2011). An application of abscisic acid (ABA) or GA had no significant effect on the activity and expression of StCDPK2 (Giammaria et al. 2011), while treatment with jasmonic acid decreased StCDPK2 expression at both these levels (Ulloa et al. 2002). The expression profile of StCDPK3 under different conditions has indicated that ABA functions as a positive and GA as a negative regulator of its expression. Agrobacterium tumefaciens-mediated transformation allowed demonstrating that StCDPK3 was expressed in early induced stolons and its expression decreased in advanced tuber formation stages (Grandellis et al. 2012). As in other species, StCDPK4 and StCDPK5 have been shown to phosphorylate and thereby activate NADPH oxidase to promote reactive oxygen species (ROS) production in response to biotic stimuli (Kobayashi et al. 2007).
The availability of the whole genome sequence of the potato (Potato Genome Sequencing Consortium 2011) allowed us to carry out a comprehensive in silico search for CDPK genes and their subsequent expression profiling. RT-qPCR expression analysis was carried out for the CDPK genes in different organs of potato. The activity and expression of the CDPKs were also determined in Solanum genotypes expressing different resistance against the pathogenic oomycete Phytophthora infestans causing late blight, the most destructive potato disease.
Materials and methods
Identification of calcium-dependent protein kinase genes in potato
Gene retrieval
All S. tuberosum genes identified in the genomic annotation as encoding a calcium-dependent protein kinase were downloaded from the public databases Spud DB (http://potato.plantbiology.msu.edu/) and Sol Genomics Network (http://solgenomics.net/). As references for the identification of CDPK genes we used GenBank NCBI sequences of potato genes described in the literature [StCDPK1 accession number (acc. no.) DQ507862, StCDPK2 acc. no. AF418563, StCDPK3 acc. no. JF308510, StCDPK4 acc. no. AB279737, StCDPK5 acc. no. AB279738]. Comparison of particular genes was performed using BLAST program (The Basic Local Alignment Search Tool). For analysis, we used 21 genes annotated as calcium-dependent protein kinases in the above-mentioned databases and two genes, StCDPK3 and StCDPK5, absent from those bases. Finally, 23 potato genes were identified as CDPKs (Table 1 and Supplementary material 1) and were subjected to further analyses. Thirty-four Arabidopsis thaliana CDPK genes used for phylogenetic analysis were downloaded from GenBank NCBI (Supplementary material 2).
Phylogenetic analysis and gene structure prediction
Full-length amino acid sequences of 23 potato and 34 A. thaliana CDPKs were aligned using ClustalX version 2.0 program with default parameters (Larkin et al. 2007). A phylogenetic tree was constructed according to the neighbor-joining method using MEGA 7.00 program (Tamura et al. 2004; Kumar et al. 2016).
The gene structures of the CDPKs were predicted by The Gene Structure Display Server (GSDS) version 2.0 (Hu et al. 2015).
Protein domains were predicted by PROSITE scan (Sigrist et al. 2013), and myristoylation sites by NMT—The MYR Predictor script (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm).
Plant material and growth conditions—in vitro cultures
Plants of Solanum tuberosum cultivar (cv) Bzura and Solanum scabrum (received from IHAR-PIB, Młochów Research Center, Poland, Plich et al. 2015) and Solanum tuberosum clone H-8105 were cultured in vitro as described previously (Polkowska-Kowalczyk et al. 2004). The plants were grown under controlled conditions: day/night 16 h/8h, fluorescent lamp 150 μmol m−2 s−1 for 16 h, day/night temperature of 22/18 °C and propagated at 4–6 week intervals.
Plant material and growth conditions—plants grown in soil
After 4 weeks of growth in vitro, Bzura plants were transferred to soil and were grown in a green-house under conditions described above. At 6–8 weeks after planting, young and mature leaves, young shoots, stolons, swollen stolons, stems, flowers, roots and tubers were harvested for analysis. The collected samples were immediately frozen in liquid nitrogen and stored at −80 °C.
Pathogen elicitor
The pathogen Phytophthora infestans (isolate MP618 from 2005, of the following race: 1.2.3.4.(5).6.7.10.11, A1 mating type and resistant to metalaxyl) received from IHAR-PIB, Młochów Research Center, Poland, was maintained on rye agar medium at 15 °C in the dark. Culture filtrate (CF), which served as an elicitor, was prepared from the pathogen grown in liquid medium. After 6 weeks of growth, the medium was separated from the oomycete, dialysed against water for 48 h and lyophilised. The CF residue was dissolved in distilled water and quantified as µg glucose equivalents ml−1, as described earlier by Polkowska-Kowalczyk et al. (2004).
Elicitor treatment of leaves
Leaves of S. tuberosum cv Bzura, S. tuberosum clone H-8105, and Solanum scabrum, which exhibited field resistance, susceptibility and non-host resistance to P. infestans, respectively, were harvested from 4-week-old plants grown in vitro. The leaves were placed on moist filter paper in Petri dishes and the culture filtrate was applied in small droplets on the abaxial surface of each leaf at a dose of 0.67 μg glucose equivalents·g−1 fresh weight (FW). As a control an equal volume of distilled water was applied. Leaves were incubated at 25 °C under continuous white light of 150 μmol m−2 s−1 from fluorescent tubes (Pila, Poland) and taken for analysis after 1, 3, 6, 18, 24, and 30 h (Polkowska-Kowalczyk et al. 2004).
RNA isolation
We used two methods for RNA isolation. Total RNA was isolated from samples (0.1 g FW) of young and mature leaves using the TRIzol reagent according to manufacturer’s protocol (Molecular Research Center, INC.). For RNA isolation from other organs, to avoid the gelling of starch and low recovery of RNA, a modified method of Kumar et al. (2007) was used. Briefly, 0.1 g FW of frozen tissue was ground with liquid nitrogen in a mortar to a fine powder. RNA was extracted with 500 µL of extraction buffer containing 100 mM Tris–HCl (pH 8), 5 mM EDTA (pH 8), 100 mM NaCl, 0.5% SDS and 1% β-mercaptoethanol. The extract was vortexed for 5 min at room temperature (RT). Next, it was centrifuged (5 min, 11,000×g) and supernatant was transferred to new tubes, 250 μL of chloroform was added and mixed for 15 min. at RT, next 250 μL of Tris-saturated phenol (pH 7.9) was added and mixed again for 15 min. The mixture was centrifuged at 4 °C for 10 min at 14,000×g and upper phase (550 μL) was transferred to a fresh tubes and extracted with an equal volume of Tris-saturated phenol (pH 7.9): chloroform: isoamyl alcohol (25:24:1; v:v:v) for 10 min. After centrifugation as above, 500 μL of upper phase was transferred to new tubes and RNA was precipitated with 50 μL of 3 M sodium acetate and 400 μL isopropanol at -80 °C for at least 1 h. RNA was pelleted by centrifugation at 14,000×g for 30 min at 4 °C. The RNA pellet was washed free of salts with 85, 80 and 75% EtOH. The residual EtOH was evaporated on ice for about 10 min and RNA was dissolved in water.
Concentration and quality of RNA were estimated using NanoDrop 2000 spectrophotometer (ThermoScientific) at 260, 280 and 230 nm. The extent of protein and carbohydrate/phenolic contamination was assessed by A260/280 and A260/230 ratios, respectively.
Reverse transcription quantitative PCR
First strand cDNA was synthesized using Enhanced Avian HS RT-PCR Kit (SIGMA-ALDRICH). After the cDNA synthesis reaction, qPCR was performed.
Primers were designed using the Primer Quest program http://eu.idtdna.com/PrimerQuest/Home/Index) (Supplementary material 3). The reaction mixture contained Real-Time 2× HS-PCR Master Mix SYBR A (A&A Biotechnology), specific forward and reverse primers (100 nM each), cDNA template (in three concentrations, each in duplicate), and water to 10 µL of final volume.
Reactions were performed in a Light Cycler LC480 (Roche) with an initial denaturation step of 95 °C for 1 min. (A&A kit) followed by 45 cycles of denaturation (95 °C for 15 s) and primer annealing-extension (60 °C for 1 min.). Fluorescence was read during the annealing-extension step of each cycle. After cycling, melting point temperature analysis was performed in the range of 60–95 °C. In each experiment background range was adjusted automatically and the cycle threshold (Ct) evaluation was adjusted manually. Quality of results was evaluated based on expected Ct differences among three cDNA amounts as well as product melting curves. Using three concentrations of cDNA permitted determining amplification efficiencies for each primer pair and normalization of all results to one cDNA concentration common for all genes. Expression of each target gene was calculated by the ΔCt method against the geometric mean of the Ct values for two reference genes (Vandesompele et al. 2002). The reference genes, elongation factor 1-α (ef1α) (acc. no. AB061263.1) and cytoplasmic ribosomal protein L2 gene (acc. no. 39816659), were used as an endogenous control to normalize variance in the quality of RNA and the amount of cDNA used. The ratio between the target and reference gene expressions was presented as the final value.
In-gel kinase assay
The in-gel kinase assay was performed according to the method described by Szczegielniak et al. (2005), with slight modifications. Briefly, 0.3 g FW of frozen Solanum leaves was ground with glass beads in liquid nitrogen in a mortar. Proteins were extracted with 100 µL of extraction buffer (50 mM Tris–HCl pH 7.5, 500 mM sucrose, 4 mM EDTA, 4 mM EGTA, 20 mM NaF, 100 mM β-glicerophosphate, 2 mM PMSF, 10 mM DTT, 40 μg/mL leupeptin, 10 μg/mL aprotinin, 4 μg/mL pepstatin, 200 μM Na3VO4). The suspension was sonicated three times for 10 s each, centrifuged at 10,280×g for 10 min at 4 °C. The supernatant was centrifuged again for 30 min at 4 °C. Protein concentration was determined using bovine serum albumin (BSA) as the standard (Bradford 1976).
Proteins (from 15 to 30 μg) from the clarified homogenate were separated on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels containing myelin basic protein (MBP; 0.25 mg/mL) as the immobilized substrate of protein kinases.
After electrophoresis, SDS was removed by washing the gel three times for 30 min each with 25 mM Tris, pH 7.5, containing 0.5 mM DTT, 0.1 mM Na3VO4, 5 mM NaF, 0.5 mg mL−1 BSA and 0.1% (v/v) Triton X-100 at room temperature. Next, the gel was equilibrated in 25 mM Tris, pH 7.5 containing 5 mM NaF, 1 mM DTT and 0.1 mM Na3VO4, for 18 h with three changes of the buffer. Then, the gels were pre-incubated for 30 min at 4 °C in 10 mL of reaction buffer (20 mM Tris, pH 7.5, 15 mM MgCl2, 2 mM DTT, and 0.33 mM Ca(CH3COOH)2 or 2 mM EGTA). Kinase activity was assayed for 1.5 h at 30 °C in the reaction buffer supplemented with 0.25 mM ATP containing 50 μCi [γ32P] ATP.
Unincorporated [γ32P] ATP was removed by washing the gels in 5% (w/v) trichloroacetic acid containing 1% (w/v) sodium phosphate. After washing, the gels were stained with Coomassie Brilliant Blue R250 (Sigma-Aldrich) and dried. The kinase activity was visualized by autoradiography or quantified using a PhosphorImager.
Results
Identification and distribution of calcium-dependent protein kinase genes in the potato genome
A genome-wide analysis of the CDPK gene family in potato (Solanum tuberosum) was performed on the complete potato genome sequence (Potato Genome Sequencing Consortium 2011) using bioinformatics methods. Only five CDPK genes had been reported earlier for S. tuberosum: StCDPK1, StCDPK2, StCDPK3, StCDPK4 and StCDPK5. Two nucleotide sequence databases were selected for an in silico search for additional genes (see Materials and methods). The first contains sequences from an international sequencing project for S. tuberosum group Phureja DM1-3 516 R44 (DM; AEWC00000000) and heterozygous diploid breeding line, S. tuberosum group Tuberosum RH89-039-16 (RH; ERP000627). The second base comprises sequences of plant genomes from the Asteraceae clade which includes the family Solanaceae. This database also contains a wealth of tools for comparative analysis of genomes.
Twenty-nine CDPK genes automatically annotated as calcium-dependent protein kinases were found in the two databases, of which only 21 genes had all the domains typical for CDPKs. Because the previously identified StCDPK3 and StCDPK5 are absent from these bases, we used for further analyses StCDPK3 (cDNA and genomic) and StCDPK5 (cDNA) sequences deposited in GenBank (NCBI). Altogether, 23 potato CDPKs were identified, designated StCPK1 - StCPK23 according to the proposed nomenclature for CDPK genes (Hrabak et al. 1996; Boudsocq and Sheen 2013) (Table 1 and Supplementary material 1). The CDPK genes are indicated by the three-letter abbreviation CPK followed by a number. The potato CDPKs identified in this study have structures typical for the CDPK family, comprising the N-terminal and C-terminal variable domains, the protein kinase domain, the autoinhibitory junction domain, and the calmodulin-like domain containing four calcium binding motifs called EF-hands (Supplementary material 4). Moreover, 14 of the potato CDPKs were found to contain predicted myristoylation sites at their N-terminus.
Among the 29 automatically annotated potato CDPKs there are eight atypical ones containing one or no calcium-binding motif. Information about these atypical kinases is included in the supplementary material (Supplementary material 5).
Phylogenetic characterization and exon-intron structure analysis
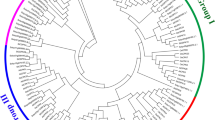
To establish the phylogenetic relationships among the StCDPK genes, a combined (S. tuberosum and A. thaliana) phylogenetic tree was constructed for the StCDPK amino acid sequences. The unrooted tree was generated from an alignment of 57 CDPK protein sequences, 23 potato and 34 A. thaliana ones. All these CDPKs could be broadly classified into four major subfamilies (subfamilies I, II, III and IV shown in Fig. 1). Subfamily I contained 11 StCDPKs: StCPK4, StCPK5, StCPK6, StCPK7, StCPK9, StCPK11, StCPK12, StCPK13, StCPK15, StCPK20, StCPK21, subfamily II 7 members: StCPK1, StCPK2, StCPK3, StCPK14, StCPK17, StCPK18, StCPK19, subfamily III 3 members: StCPK8, StCPK10, StCPK23, and subfamily IV only 2 StCDPKs: StCPK16 and StCPK22.
According to Boudet et al. (2001), the intron-exon organization and the type of the introns reflect the evolutionary history of some gene families. The potato CDPKs were examined in this respect to obtain further insight into their evolution. Each of the previously identified four subfamilies showed similar intron-exon organization of genes within the family and markedly different patterns between them (Supplementary material 6). Most of the 11 StCDPKs from the first subfamily had six or seven introns, all the genes from the second and third subfamilies had seven or eight introns, and those from the fourth subfamily had the highest number of introns, eleven. Since for StCPK5 from the first subfamily only the cDNA sequence is available, its exon-intron organization could not be determined.
Chromosomal distribution of potato CDPK genes
To establish the chromosomal locations of the encoding genes, nucleotide sequences of each StCDPK were used to search the potato genome database using BLASTN. The genes were found on 11 out of the potato 12 chromosomes (except chromosome 9). Five chromosomes had only a single CDPK gene each: chromosome 2—StCPK22; chromosome 3—StCPK16; chromosome 4—StCPK14; chromosome 7—StCPK2; chromosome 8—StCPK19. Four chromosomes carried two CDPK genes each: chromosome 5—StCPK6 and StCPK7; chromosome 6—StCPK11 and StCPK23; chromosome 11—StCPK15 and StCPK17; chromosome 12—StCPK1 and StCPK18. Three CDPK genes were localized on chromosome 1—StCPK8, StCPK9 and StCPK20, and five genes: StCPK4, StCPK10, StCPK12, StCPK13 and StCPK21 on chromosome 10 (Table 1). On some chromosomes the StCDPKs form clusters, which suggests recent gene duplication. Such a differential genomic distribution of the StCDPKs suggests gradual expansion of this gene family.
Organ-specific expression of potato CDPK genes
Various potato organs obtained from S. tuberosum cv Bzura, such as young and mature leaves, young shoots, stems, roots, stolons, swollen stolons, flowers and tubers were used for expression profiling of CDPKs by RT-qPCR. Transcripts of all 23 CDPKs were present in all the samples analysed, although their level varied greatly (Fig. 2 and Supplementary material 7). The organs differed from one another not only in the overall level but also in the spectrum of the expressed CDPKs. Highly similar patterns of expression were observed for two pairs of organs: stolons and young shoots, and young and mature leaves, respectively. The highest level of overall StCDPKs expression was found in young shoots and stolons, mainly of StCPK2, 4, 5, 12, 15, 16, 17, and 22. Roots and flowers showed a medium level of CDPKs expression, while in stems, tubers, swollen stolons and leaves (young and mature), the expression was the lowest and generally similar in all these organs (Fig. 2).
Expression of individual potato CPK genes in the indicated organs. Please note different ordinate scales for different organs. Expression profiles are represented by at least three independent experiments each in two replicates. The data are the mean values ± SD (n ≥ 6). Relative expression is presented in relation to reference genes
StCPK6, 7, 13 and 18 were expressed mostly in flowers, StCPK4, 5, 12, 15, 17 and 22 in young shoots, roots and stolons. Transcripts of StCPK1, 13, 14, and 19 were hardly detectable in some of the organs analysed and were sometimes even below the limits of detection (Supplementary material 7). It seems therefore likely that some CDPKs may be expressed only in response to certain stimuli, at specific developmental stages, and/or in certain cell types.
Summing up, the potato CDPKs exhibit quite complex organ-specific expression patterns, suggesting that some function in defined tissues or developmental stages, while others have broader functions.
Expression of CDPKs in Solanum leaves genotypes treated with elicitor from P. infestans
To learn more about the potential function of the CDPKs in plant disease resistance, kinetics of all the CDPK genes was investigated in potato leaves treated with an elicitor—a culture filtrate (CF) of P. infestans. For comparison we used leaves from three Solanum genotypes showing substantially different susceptibility to the pathogen: S. tuberosum cv Bzura, S. tuberosum clone H-8105 and Solanum scabrum exhibiting respectively field resistance, susceptibility and non-host resistance to P. infestans. Changes in expression of 15 CDPKs were observed after the elicitor treatment: five genes from the first subfamily (StCPK4,5,6,7,15), six from the second subfamily (StCPK1,2,3,14,17,18), three from the third subfamily (StCPK8,10,23) and one from the fourth subfamily (StCPK16) (Fig. 3). Interestingly, in the non-host S. scabrum an increase in gene expression occurred between 0.5 and 6 h after CF treatment, reaching for StCPK23 ca. 315% compared to water-treated control (100%) (Supplementary material 8). In the susceptible H-8105, also StCPK23 was the most strongly induced (430% of control) but only at prolonged treatment (18–30 h). In the field resistant Bzura leaves, the maximal transcript level was noticed between 6 and 18 h after the elicitor treatment, and the increase was the highest for StCPK4 (264% of control).
Expression of various CDPK genes in elicitor-treated leaves of three Solanum genotypes expressed as % of control levels. Transcript levels at time-points indicated are expressed relative to control value determined as the average of all time points in control (water-treated) leaves. At least two independent biological series of each experiment were performed. Data are mean values ± SD (n ≥ 4)
Activity of CDPKs after treatment with elicitor from P. infestans
To check the influence of the P. infestans elicitor on the CDPK kinase activity, we determined the enzymatic activity using the in-gel kinase assay in the presence of calcium ions and, as a negative control, without calcium ions. The activity was studied in leaf extracts prepared from the three different Solanum genotypes treated with the elicitor from P. infestans (CF) for between 0 and 24 h. In all the genotypes studied the activity of CDPKs increased in response to the CF treatment, but with different kinetics and maximal intensity (Fig. 4). In the resistant Bzura genotype the CDPKs activity rose gradually between 0.5 and 3 h of treatment to reach ca. 155% of the control level and remained elevated (ca. 130–150%) until the end of the experiment (24 h). In contrast, in the susceptible clone H-8105, after an early increase to about 130% of control, the CDPKs activity returned to the control level after 6 h and remained at that level. In non-host S. scabrum the CDPKs activity increased gradually to reach a maximum at 6 h of CF treatment (193% of control), remained elevated until the 18th hour and then dropped rapidly to the control level.
Activity of CDPK in elicitor-treated leaves of three Solanum genotypes shown as % of control levels at various time points. “Zero” denote average values represented by control leaves treated with water from 0 to 24 h. At least two independent biological series of experiments were performed. Data are mean values ± SD (n ≥ 4)
Thus, in the resistant genotypes a progressive, marked and long-lasting increase of the kinase activity was observed while the susceptible genotype only showed early, weak and transient activation of the CDPKs. These results suggest a correlation between CDPK activation and resistance of potato against P. infestans.
Discussion
Potato is one of a major crops sustaining the world’s population owing to the high nutritional value of its underground tubers. Understanding the signalling pathways that regulate its development and stress responses is key to improving the potato yield, disease resistance, storability and dietary properties.
Calcium-dependent protein kinases (CDPKs), one of the largest groups of plant calcium sensors, bind calcium ions directly which modulates their activity. The calcium-activated kinases regulate diverse developmental processes and defence responses of plants.
The present comprehensive study of CDPK genes in the potato was made possible by the availability of the complete genomic sequence (Potato Genome Sequencing Consortium 2011) and provides a foundation for further functional investigation of this important gene family in Solanaceae. The potato genome encodes at least 23 CDPKs similar to the 20–40 CDPK genes present in most other plants. The expansion of gene families in plant genomes is believed to have occurred by various mechanisms such as genome-wide, tandem and dispersed duplications. The potato genome appears to exhibit high plasticity, as gene loss or duplication events and other structural mutations are found with high frequency (Potato Genome Sequencing Consortium 2011).
The domain organization of the 23 potato CDPKs is typical for the CDPKs from other plant species. All potato CDPKs have four EF-hands, and 14 of them contain myristoylation motifs indicating that they undergo the modification promoting protein-membrane and protein-protein interactions (Xu et al. 2015). In potato, like in rice, maize and Arabidopsis thaliana, some of the CDPK-like protein kinases (8 in potato) contain less than four, or even none, EF-hands. In the potato genome the 23 CDPKs genes are distributed apparently randomly among chromosomes. In the tomato 29 SlCDPKs genes are distributed in all chromosomes also highly unevenly, which the authors interpreted as indicating evolution events and recent expansion of the family (Wang et al. 2015).
CDPKs are involved in various physiological processes at different stages of plant development (Liu et al. 2016). To investigate the expression profiles of the CDPK genes in potato development, we determined their transcript levels in different organs such as young and mature leaves, young shoots, stems, roots, stolons, swollen stolons, flowers and tubers. Although transcripts of at least some CDPKs were detected in all the organs, their expression levels varied greatly between individual genes and also between organs. Only in two pairs of organs, young and mature leaves, and stolons and young shoots, the expression patterns were highly similar. Notably, young shoots, roots and stolons showed particularly high expression of CDPKs mostly from the first subfamily (StCPK4, 5, 12, 15) and StCPK17 from the second subfamily.
Interestingly, StCPK3 is highly similar in its amino acid sequence to tobacco NtCDPK1 and tomato LeCPK1 (Yoon et al. 1999; Rutschmann et al. 2002). These three genes are expressed at a very high level in roots. A down-regulation of NtCDPK1 in transgenic tobacco resulted in smaller plants showing abnormal root development, with reduced lateral root formation and impaired elongation (Lee et al. 2006). By analogy, StCPK3 likely participates in the development of main and lateral roots (Grandellis et al. 2012).
Four StCPKs: StCPK6, 7, 13 and 18, were expressed exclusively in flowers, indicating their possible role in flower development. This is in concert with earlier results. According to Schulz et al. (2013), CDPKs have a major role in pollen tube growth. Twelve members of the Arabidopsis CDPK gene family are predominantly expressed in pollen (Myers et al. 2009). For AtCPK2/AtCPK20 and AtCPK17/AtCPK34, a role in pollen tube growth has been demonstrated experimentally in double mutant lines (Gutermuth et al. 2013). StCPK13, expressed exclusively in potato flowers, is very similar to AtCPK2/AtCPK20, while StCPK18, highly expressed in potato flowers, is closely related to AtCPK17/AtCPK34. AtCPK2/CPK20 and AtCPK17/AtCPK34 are involved in the regulation of pollen tube growth, and AtCPK2/CPK20 influence the anion channel SLAH3 (Gutermuth et al. 2013). Moreover, PiCDPK1 from petunia (Petunia inflata), similar to AtCPK17/AtCPK34, has also been implicated in pollen tube growth polarity (Yoon et al. 2006). One of the maize CDPK genes (designated by us as ZmCPK7 according to the nomenclature by Ma et al. 2013), also similar to AtCPK17/AtCPK34, is expressed specifically during late stages of pollen development. Specific inhibition of the ZmCPK7 expression by antisense oligonucleotide resulted in impaired germination and growth of pollen tube (Estruch et al. 1994). According to Ray et al. (2007), OsCPK2 and 14 from rice that show homology to PiCDPK1 and AtCPK17 and 34, and OsCPK25 and 26, highly similar to ZmCPK7, are expressed during late panicle development and thus could be involved in a similar function during pollen maturation and/or pollen tube growth.
A process typical for potato is tuberization that results in differentiation of a specialized shoot (stolon) into a storage organ (the tuber). Tuber development is a tightly controlled process triggered by both external and internal factors; among them calcium ions and protein kinases play an important role (Balamani et al. 1986; Raíces et al. 2003). It has been suggested that sequential activation of specific CDPKs with distinct biochemical properties and subcellular localization could be essential for the co-ordination of multiple Ca2+ signals triggered upon tuberization (Raíces et al. 2003). StCDPK1 and StCDPK3, with different substrate specificities and cellular distribution, were found associated with early and induced stolons, respectively. StCDPK1 was expressed in tuberizing stolons and sprouting tubers (Raíces et al. 2001; Gargantini et al. 2009), whereas expression of StCDPK3 was abundant in stolons, roots and leaves (Grandellis et al. 2012). Transcript of another CDPK, StCDPK2, was present in all plant tissues studied and during different developmental processes - sprouting and tuberization (Giammaria et al. 2011); its level was the highest in actively growing young leaves. In our experimental system, three CDPKs described before were expressed in all organs analysed, although expression of StCPK1 was very low. StCPK2 and StCPK3 were mainly expressed in roots, stolons, young shoots, and flowers. StCPK4, 5, 12, 15, 16, and 17 were highly expressed in stolons and were the major ones in a rapidly growing organ—the young shoots. The differences in expression of individual CDPKs likely correlate with their participation in specific signal transduction pathways in different organs.
Accumulating evidence indicates that CDPKs play an important role in plant immunity. For example, Fu et al. (2013) demonstrated that overexpression of constitutively active OsCPK10 enhanced Arabidopsis resistance against Pseudomonas syringae pv. tomato. Our present study indicated a likely involvement of CDPKs in Solanum species in response to an elicitor, a culture filtrate (CF) from the pathogenic oomycete Phytophthora infestans causing late blight, the most destructive potato disease. We determined expression profiles of all the potato CDPKs and CDPK activities in three Solanum genotypes expressing different types and level of resistance against P. infestans. Expression of all 15 CDPKs and the kinase activity increased after CF treatment in all these genotypes, but with strikingly different patterns in the sensitive and the resistant ones. These results are in agreement with the earlier findings of Vleeshouwers et al. (2000). Their cytological study indicated that the severity and timing of defence response varied depending on the form and level of resistance exhibited by a Solanum genotype. Those authors suggested that in the Solanum/P. infestans interaction resistance is a quantitative rather than a qualitative trait. In our experimental system the magnitude and duration of gene induction and activity upregulation of the CDPKs correlated positively with the level of resistance of the Solanum genotypes to P. infestans.
Some authors have documented a relation between CDPKs and production of reactive oxygen species (ROS). Thus, Kobayashi et al. (2007) observed phosphorylation of NADPH oxidase by StCDPK4 and StCDPK5 that resulted in oxidative burst. We observed a positive correlation between increased activity and gene expression of CDPKs (present results) and ROS generation (Polkowska-Kowalczyk et al. 2004) in resistant genotypes in response to P. infestans, but not such correlation was found for the susceptible H-8105. Taken together, the earlier and present results concerning different Solanum genotypes (Polkowska-Kowalczyk et al. 2004, 2011) indicate that an effective defence against P. infestans involves cooperation among the diverse elicitor-induced processes.
In conclusion, the potato genome contains at least 23 members of the CDPK gene family. Most of these genes exhibit markedly different expression levels in various organs, suggesting that they play specific different roles in potato development. Increased CDPK expression and activity in response to the elicitor from P. infestans indicates involvement of CDPKs in the defence against this pathogen. Due to the high economic significance of the potato, it is imperative to study defence mechanisms occurring in response to pathogen attack.
References
Asano T, Tanaka N, Yang G, Hayashi N, Komatsu S (2005) Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol 46(2):356–366
Baillie BK, Belda-Baillie CA, Maruyama T (2000) A calcium-dependent protein kinase functions in wound healing in Ventricaria ventricosa (chlorophyta). J Phycol 36:1145–1152
Balamani V, Veluthambi K, Poovaiah BW (1986) Effect of calcium on tuberization in potato (Solanum tuberosum L.). Plant Physiol 80:856–858
Billker O, Lourido S, Sibley LD (2009) Calcium-dependent signalling and kinases in apicomplexan parasites. Cell Host Microbe 5:612–622
Boudet N, Aubourg S, Toffano-Nioche C, Kreis M, Lecharny A (2001) Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res 11:2101–2114
Boudsocq M, Sheen J (2013) CDPKs in immune and stress signalling. Trends Plant Sci 18:30–40
Boudsocq M, Willmann MR, McCarmack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464:418–422
Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Broad Institute of Harvard and MIT (2010) Phytophthora infestans Sequencing Project. https://www.broadinstitute.org
Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B (2014) Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol 165:688–704
Cheng S-H, Willmann MR, Chen H-C, Sheen J (2002) Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129:469–485
Coca M, San Segundo B (2010) AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J 63:526–540
Dubrovina AS, Kiselev KV, Khristenko VS, Aleynova OA (2017) The calcium-dependent protein kinase gene VaCPK29 is involved in grapevine responses to heat and osmotic stresses. Plant Growth Regul 82:79–89
Estruch JJ, Kadwell S, Merlin E, Crossland L (1994) Cloning and characterization of a maize pollen-specific calcium-dependent calmodulin-independent protein kinase. Proc Natl Acad Sci USA 91:8837–8841
Fu L, Yu X, An Ch (2013) Overexpression of constitutively active OsCPK10 increases Arabidopsis resistance against Pseudomonas syringae pv. tomato and rice resistance against Magnaporthe grisea. Plant Physiol Biochem 73:202–210
Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J (2013) Bifurcation of Arabidopsis NLR immune signalling via Ca2+-dependent protein kinases. PloS Pathol 9:e1003127
Gargantini PR, Giammaria V, Grandellis C, Feingold SE, Maldonado S, Ulloa RM (2009) Genomic and functional characterization of StCDPK1. Plant Mol Biol 70:153–172
Giammaria V, Grandellis C, Bachmann S, Gargantini PR, Feingold SE, Bryan G, Ulloa RM (2011) StCDPK2 expression and activity reveal a highly responsive potato calcium-dependent protein kinase involved in light signalling. Planta 233:593–609
Grandellis C, Giammaria V, Bialer M, Santin F, Lin T, Hannapel DJ, Ulloa RM (2012) The novel Solanum tuberosum calcium dependent protein kinase, StCDPK3, is expressed in actively organs. Planta 236:1831–1848
Gutermuth T, Lassig R, Portes M-T, Maierhofer T, Romeis T, Borst JW, Hedrich R, Feijó JA, Konrad KR (2013) Pollen tube growth regulation by free anions depends on the interaction between the anion channel SLAH3 and calcium-dependent protein kinases CPK2 and CPK20. Plant Cell 25:4525–4543
Harmon AC, Gribskov M, Gubrium E, Harper JF (2001) The CDPK superfamily of protein kinases. New Phytol 151:175–183
Hrabak EM, Dickmann LJ, Satterlee JS, Sussman MR (1996) Characterization of eight new members of the calmodulin-like domain protein kinase gene family of Arabidopsis thaliana. Plant Mol Biol 31:405–412
Hrabak EM, Chan CWM, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu JK, Harmon AC (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132:666–680
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Hu Z, Lv X, Xia X, Zhou J, Shi K, Yu J, Zhou Y (2016) Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front Plant Sci 7:469. https://doi.org/10.3389/fpls.2016.00469
Ivashuta S, Liu J, Liu J, Lohar DP, Haridas S, Bucciarelli B, VandenBosch KA, Vance CP, Harrison MJ, Gantt JS (2005) RNA interference identiWes a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell 17:2911–2921
Kamiyoshihara Y, Iwata M, Fukaya T, Tatsuki M, Mori H (2010) Turnover of LeACS2, a wound-inducible 1-aminocyclopropane-1-carboxylic acid synthase in tomato, is regulated by phosphorylation/dephosphorylation. Plant J 64:140–150
Klimecka M, Muszyńska G (2007) Structure and functions of plants calcium-dependent protein kinases. Acta Biochim Polon 54(2):219–233
Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19:1065–1080
Kong X, Lv W, Jiang S, Zhang D, Cai G, Pan J, Li D (2013) Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genom 14:433. https://doi.org/10.1186/1471-2164-14-433
Kumar GNM, Iyer S, Knowles NR (2007) Extraction of RNA from fresh, frozen, and lyophilized tuber and root tissues. J Agric Food Chem 55:1674–1678
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWiliam H, Valentiv F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948
Lee SS, Yoon GM, Rho EJ, Moon E, Pai HS (2006) Functional characterization of NtCDPK1 in tobacco. Mol Cells 28:141–146
Li AL, Zhu YF, Tan XM, Wang X, Wei B, Guo HZ, Zhang ZL, Chen XB, Zhao GY, Kong XY (2008) Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol Biol 66:429–443
Liu W, Li W, He Q, Khan Daud M, Chen J, Zhu S (2014) Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS ONE 9:e98189. https://doi.org/10.1371/journal.pone.0098189
Liu H, Che Z, Zeng X, Zhou X, Sitoe HM, Wang H, Yu D (2016) Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Funct Integr Genom 16:481–493
Lu SX, Hrabak EM (2002) An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol 128:1008–1021
Ma P, Liu J, Yang X, Ma R (2013) Genome-wide identification of the maize calcium-dependent protein kinase gene family. Appl Biochem Biotechnol 169:2111–2125
McCurdy DW, Harmon AC (1992) Calcium-dependent protein kinase in the green alga Chara. Planta 188:54–61
Myers C, Romanowsky SM, Barron YD, Garg S, Azuse CL, Curran A, Davis RM, Hatton J, Harmon AC, Harper JF (2009) Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J 59:528–539
Plich J, Tatarowska B, Lebecka R, Śliwka J, Zimnoch-Guzowska E, Flis B (2015) R2-like gene contributes to resistance to Phytophthora infestans in polish potato cultivar Bzura. Am J Potato Res 92:350–358
Polkowska-Kowalczyk L, Wielgat B, Maciejewska U (2004) The elicitor-induced oxidative processes in leaves of Solanum species with differential polygenic resistance to Phytophthora infestans. J Plant Physiol 161:913–920
Polkowska-Kowalczyk L, Wielgat B, Maciejewska U (2011) Involvement of phospholipase A2 in the response of Solanum species to an elicitor from Phytophthora infestans. Acta Physiol Plant 33:2521–2531
Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–197
Raíces M, Chico JM, Téllez-Iňón MT, Ulloa RM (2001) Molecular characterization of StCDPK1, a calcium-dependent protein kinase from Solanum tuberosum that is induced at the onset of tuber development. Plant Mol Biol 46:591–601
Raíces M, Gargantini PR, Chinchilla D, Crespi M, Téllez-Iňón MT, Ulloa RM (2003) Regulation of CDPK isoforms during tuber development. Plant Mol Biol 52:1011–1024
Ray S, Agarwal P, Arora R, Kapoor S, Tyagi AK (2007) Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol Genet Genom 278:493–505
Romeis T, Herde M (2014) From local to global: CDPKs in systemic defence signalling upon microbial and herbivore attack. Curr Opin Plant Biol 20:1–10
Romeis T, Ludwig AA, Martin R, Jones JD (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20:5556–5567
Rutschmann F, Stalder U, Piotrowski M, Oecking C, Schaller A (2002) LeCPK1, a calcium-dependent protein kinase from tomato: plasma membrane targeting and biochemical characterization. Plant Physiol 129:156–168
Schulz P, Herde M, Romeis T (2013) Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol 163:523–530
Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I (2013) New and continuing developments at PROSITE. Nucleic Acids Res 41:D344-D347. https://doi.org/10.1093/nar/gks1067
Szczegielniak J, Klimecka M, Liwosz A, Ciesielski A, Kaczanowski S, Dobrowolska G, Harmon AC, Muszyńska G (2005) A wound-responsive and phospholipid-regulated maize calcium-dependent protein kinase. Plant Physiol 139:1970–1983
Szczegielniak J, Borkiewicz L, Szurmak B, Lewandowska-Gnatowska E, Statkiewicz M, Klimecka M, Ciesla J, Muszynska G (2012) Maize calcium-dependent protein kinase (ZmCPK11): local and systemic response to wounding, regulation by touch and components of jasmonate signaling. Physiol Plant 146:1–14
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035
Ulloa RM, Raíces M, MacIntosh GC, Maldonado S, Téllez-Iňón MT (2002) Jasmonic acid affects plant morphology and calcium-dependent protein kinase expression and activity in Solanum tuberosum. Physiol Plant 115:417–427
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. https://doi.org/10.1186/gb-2002-3-7-research0034
Vleeshouwers VGAA, van Dooijweert W, Govers F, Kamoun S, Colon LT (2000) The hypersensitive response is acciociated with host and nonhost resistance to Phytophthora infestans. Planta 210:853–864
Wang J-P, Xu Y-P, Munyampundu J-P, Liu T-Y, Cai X-Z (2015) Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: genome-wide identification and functional analyses in disease resistance. Mol Genet Genom. https://doi.org/10.1007/s00438-015-1137-0
Xu X, Liu M, Lu L, He M, Qu W, Xu Q, Qi X, Chen X (2015) Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol Genet Genom 290:1403–1414
Yoon GM, Cho HS, Ha HJ, Liu JR, Lee HS (1999) Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol Biol 39:991–1001
Yoon GM, Dowd PE, Gilroy S, McCubbin AG (2006) Calcium-dependent protein kinase isoforms in Petunia have distinct functions in pollen tube growth, including regulating polarity. Plant Cell 18:867–878
Zhang XS, Choi JH (2001) Molecular evolution of calmodulin-like domain protein kinases (CDPKs) in plants and protists. J Mol Evol 53:214–224
Zhang K, Han Y-T, Zhao F-L, Hu Y, Gao Y-R, Ma Y-F, Zheng Y, Wang Y-J, Wen Y-Q (2015) Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol 15:164. https://doi.org/10.1186/s12870-015-0552-z
Zuo R, Hu R, Chai G, Xu M, Qi G, Kong Y, Zhou G (2013) Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Mol Biol Rep 40:2645–2662
Acknowledgements
We wish to thank Prof. Grażyna Dobrowolska for helpful discussion and Prof. Jan Fronk for critical reading of the manuscript. The authors are also grateful to Irena Dzikowska for technical support in plant cultures. We are grateful to IHAR-PIB, Młochów Research Center, Poland for providing the isolate of P. infestans (isolate MP618) and Solanum genotypes.
Funding
This work was supported by the National Science Centre, Poland, project 2012/05/B/NZ3/00911 and partially by the Ministry of Science and Higher Education, Poland, Project 0329/B/P01/2008/34.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10725_2017_341_MOESM4_ESM.tif
Domain structure of protein kinases in the potato CDPK family. Orange bars represents the catalytic serine/threonine domain, black bars the autoinhibitory junction domain, and blue bars the EF-hand motifs of the calcium-binding domain. Solid lines at N and C termini indicate regions of variable length. Every blue bar consists of two EF-hands. In StCPK11 two blue bars overlaps (TIF 11697 KB)
10725_2017_341_MOESM6_ESM.tif
Exon-intron structure of potato CDPK genes. Left-hand panel shows phylogenetic tree of S. tuberosum CPKs genes. Numbers on branches indicate the bootstrap percentage values calculated from 1000 replicates, and values below 50 % are not shown. Right-hand panel illustrates gene structures as determined by comparison of cDNA and genomic sequences using GSDS 2.0. Exons (thick lines) and introns (thin lines) are drawn to scale. Numbers indicate intron phase. For StCPK5 no genomic sequence is available (TIF 33393 KB)
10725_2017_341_MOESM7_ESM.tif
Organ-specific expression of individual potato CPK genes. Results from Fig. 2 were grouped according to the gene and drawn in a common scale. Expression profiles are represented by at least three independent experiments each in two replicates. The data are the mean values ± SD (n ≥ 6). Relative expression is presented in relation to reference genes (TIF 5330 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gromadka, R., Cieśla, J., Olszak, K. et al. Genome-wide analysis and expression profiling of calcium-dependent protein kinases in potato (Solanum tuberosum). Plant Growth Regul 84, 303–315 (2018). https://doi.org/10.1007/s10725-017-0341-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-017-0341-9