Abstract
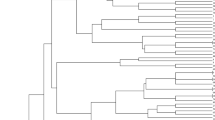
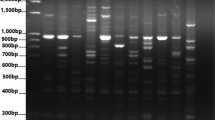
Ancient species (Triticum monococcum and Triticum dicoccum) of wheat represent a valuable genetic resource for breeding and genetic research. In this study, 81 selected genotypes and 7 commercial cultivars were evaluated with 11 simple sequence repeat (SSR) molecular markers. A total of 93 SSR alleles were detected, giving an average of 8.45 alleles per locus. Consequently, a total of 88 genotypes were assessed for their mean expected heterozygosity (He = 0.486), observed heterozygosity (Ho = 0.121), polymorphism information content (PIC = 0.68), and Shannon’s information index (I = 0.918). The clustering analysis separated the genotypes into five subclusters based on the genetic similarity coefficient. Analysis of molecular variance (AMOVA) was performed to evaluate five different clusters. The result of AMOVA was defined as genetic deviation from expectation for clusters (Fis = 0.877, Fit = 0.903, and Fst = 0.211). In Nei’s pairwise genetic identity, the highest and lowest were observed between P2-P5 populations (0.39) and P1-P2 populations (0.838). The wide variety of wheat lines can be used as a genetic resource in designing a wheat breeding program to develop new cultivars adapted to different geographic and climatic conditions and can also contribute to breeding programs around the world.



Similar content being viewed by others
Data availability
All data needed to conduct this study is provided within the manuscript. Sample spike pictures for einkorn wheat and emmer wheat are included in the supplement.
References
Asmamaw M, Keneni G, Tesfaye K (2019) Genetic diversity of ethiopian durum wheat (Triticum durum Desf.) Landrace collections as reveled by SSR markers. Adv Crop Sci Technol 7:1–9. https://doi.org/10.4172/2329-8863.1000413
Aydin A, Ince AG, Uygur Gocer E, Karaca M (2018) Single cotton seed DNA extraction without the use of enzymes and liquid nitrogen. Fresenius Env Bulletin 27:6722–6726
Bhandawat A, Sharma H, Pundir N, Madhawan A, Roy J (2020) Genome-wide identification and characterization of novel non-coding RNA-derived SSRs in wheat. Mol Biol Rep 47:6111–6125. https://doi.org/10.1007/s11033-020-05687-x
Bulunuz Palaz E, Demirel F, Adali S, Demirel S, Yilmaz A (2023) Genetic relationships of salep orchid species and gene flow among Serapias vomeracea× anacamptis morio hybrids. Plant Biotechnol Report 17:315–327. https://doi.org/10.1007/s11816-022-00782-w
Celik I, Aydin A (2023) SSR mining of black cumin (Nigella sativa L.) transcriptome for molecular marker development. Genet Res Crop Evol 70:629–638. https://doi.org/10.1007/s10722-022-01451-1
Chao S, Zhang W, Dubcovsky J, Sorrells M (2007) Evaluation of genetic diversity and genome-wide linkage disequilibrium among US wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci 47:1018–1030. https://doi.org/10.2135/cropsci2006.06.0434
Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philosophical Trans Royal Society B Biol Sci 363:557–572. https://doi.org/10.1098/rstb.2007.2170
Demirel F (2020) Genetic diversity of emmer wheats using iPBS markers. Europ J Sci Technolo 1:640–646. https://doi.org/10.31590/ejosat.814537
Demirel F, Eren B (2020) Production projection of einkorn and emmer wheat cultivated in Turkey. J Agri 3:1–5. https://doi.org/10.46876/ja.716882
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:39–40
Earl DA, vonHoldt BM (2012) Structure Harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Res 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Ellegren H, Moore S, Robinson N, Byrne K, Ward W, Sheldon BC (1997) Microsatellite evolution–a reciprocal study of repeat lengths at homologous loci in cattle and sheep. Mol Biol Evol 14:854–860. https://doi.org/10.1093/oxfordjournals.molbev.a025826
Eujayl I, Sorrells ME, Baum M, Wolters P, Powell W (2002) Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet 104:399–407. https://doi.org/10.1007/s001220100738
Gupta PK, Varshney RK, Prasad M (2002) Molecular markers: principles and methodology. In: Jain SM, Brar DS, Ahloowalia BS (eds) Molecular techniques in crop improvement. Springer, Dordrecht, pp 9–54
Gupta PK, Langridge P, Mir RR (2010) Marker-assisted wheat breeding: present status and future possibilities. Mol Breed 26:145–161. https://doi.org/10.1007/s11032-009-9359-7
Gurcan K, Demirel F, Tekin M, Demirel S, Akar T (2017) Molecular and agro-morphological characterization of ancient wheat landraces of turkey. BMC Plant Biol 17:171. https://doi.org/10.1186/s12870-017-1133-0
Harlan JR (1998) The living fields: our agricultural heritage. Cambridge University Press, UK
Hosking LK, Boyd PR, Xu CF, Nissum M, Cantone K, Purvis IJ, Khakhar R, Barnes MR, Liberwirth U, Hagen Mann K, Ehm MG, Riley JH (2002) Linkage disequilibrium mapping identifies a 390 kb region associated with CYP2D6 poor drug metabolising activity. Pharmacogenomics J 2:165–175. https://doi.org/10.1038/sj.tpj.6500096
Kabbaj H, Sall AT, Al-Abdallat A, Geleta M, Amri A, Filali-Maltouf A, Belkadi B, Ortiz R, Bassi FM (2017) Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of alleles exchange. Fronti Plant Sci 8:1277. https://doi.org/10.3389/fpls.2017.01277
Kamara MM, Rehan M, Ibrahim KM, Alsohim AS, Elsharkawy MM, Kheir AM, Hafez EM, El-Esawi MA (2020) Genetic diversity and combining ability of white maize inbred lines under different plant densities. Plants 9:1140. https://doi.org/10.3390/plants9091140
Karakaya O, Yaman M, Balta F, Yilmaz M, Balta MF (2023) Assessment of genetic diversity revealed by morphological traits and ISSR markers in hazelnut germplasm (Corylus avellana L.) from Eastern Black Sea Region, Turkey. Genet Res Crop Evol 70:525–537. https://doi.org/10.1007/s10722-022-01444-0
Kumar S, Stecher G, Tamura K (2016a) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Kumar P, Yadava RK, Kumar S, Kumar P (2016b) Molecular diversity analysis in wheat genotypes using SSR markers. Elect J Plant Breed 7:464–468. http://dx.doi.org/10.5958/0975-928X.2016b.00060.0
Liu BH (1997) Statistical genomics: linkage, mapping and QTL Analysis. CRS Press, Boca Raton, pp 10–24
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Matsuoka Y (2011) Evaluation of polyploid Triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid specification in their diversification. Plant Cell Physiol 52:750–764. https://doi.org/10.1093/pcp/pcr018
Munaweera TIK, Jayawardana NU, Rajaratnam R, Dissanayake N (2022) Modern plant biotechnology as a strategy in addressing climate change and attaining food security. Agr Food Security 11:1–28. https://doi.org/10.1186/s40066-022-00369-2
Pachauri A, Sarawgi AK, Bhandarkar S, Ojha GC (2017) Genetic variability and association study for yield contributing traits of promising core rice germplasm accessions (Oryza sativa L). Res Crops 18:133–138. https://doi.org/10.5958/2348-7542.2017.00023.7
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Peng JH, Bai Y, Haley SD, Lapitan NLV (2009) Microsatellite-based molecular diversity of bread wheat germplasm and association mapping of wheat resistance to the russian wheat aphid. Genetica 135:95–122. https://doi.org/10.1007/s10709-008-9262-x
Pinar H, Uzun A, Unlu M, Yaman M (2019) Genetic diversity in turkish banana (Musa cavendishii) genotypes with DAMD markers. Fresen Env Bulletin 28:459–463
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Rafalski JA, Tingey SV (1993) Genetic diagnostics in plant breeding: RAPDs, microsatellites and machines. Trend Genet 9:275–280. https://doi.org/10.1016/0168-9525(93)90013-8
Ramya P, Jain N, Singh GP, Singh PK, Prabhu KV (2015) Population structure, molecular and physiological characterisation of elite wheat varieties used as parents in drought and heat stress breeding in India. Ind J Genet Pl Breed 75:250–252. https://doi.org/10.5958/0975-6906.2015.00038.3
Salem KF, Röder MS, Börner A (2015) Assessing genetic diversity of egyptian hexaploid wheat (Triticum aestivum L.) using microsatellite markers. Genet Res Crop Evol 62:377–385. https://doi.org/10.1007/s10722-014-0159-5
Salunkhe A, Tamhankar S, Tetali S, Zaharieva M, Bonnett D, Trethowan R, Misra S (2013) Molecular genetic diversity analysis in emmer wheat (Triticum dicoccon Schrank) from India. Genet Res Crop Evol 60:165–174. https://doi.org/10.1007/s10722-012-9823-9
Sevindik E, Efe F (2021) Molecular genetic diversity and phylogenetic analyses of Punica granatum L. populations revealed by ISSR markers and chloroplast (cpDNA) trnL-F region. Erwerbs-Obstbau 63:339–345. https://doi.org/10.1007/s10341-021-00581-7
Sönmezoglu ÖA, Terzi B (2018) Characterization of some bread wheat genotypes using molecular markers for drought tolerance. Physiol Mol Biol Plant 24:159–166. https://doi.org/10.1007/s12298-017-0492-1
Soriano JM, Villegas D, Aranzana MJ, García del Moral LF, Royo C (2016) Genetic structure of modern durum wheat cultivars and Mediterranean landraces matches with their agronomic performance. PLoS ONE 11:e0160983. https://doi.org/10.1371/journal.pone.0160983
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceed Nation Acad Sci 101:11030–11035. https://doi.org/10.1073/pnas.0404206101
Teklu Y, Hammer K, Huang XQ, Röder MS (2006) Analysis of microsatellite diversity in Ethiopian tetraploid wheat landraces. Genet Res Crop Evol 53:1115–1126. https://doi.org/10.1007/s10722-005-1146-7
Uzun A, Pinar H, Yaman M, Yigit MA, Cakiroglu Y, Karakaya A, Uysal M, Ozturk G, Yilmaz KU, Gurcan K, Ercisli S (2022) Identification of genetic diversity in wild pear (Pyrus elaeagrifolia Pall.) Genotypes collected from different regions of turkey with SSR marker system. Genetika 54:109–118. https://doi.org/10.2298/GENSR2201109U
Vaiman D, Mercier D, Moazami-Goudarzi K, Eggen A, Ciampolini R, Lépingle A, Velmala R, Kaukinen J, Varvio SL, Martin P, Levéziel H, Guérin G (1994) A set of 99 cattle microsatellites: characterization, synteny mapping, and polymorphism. Mamm Genome 5:288–297. https://doi.org/10.1007/BF00389543
Wang S, Wong D, Forrest K, Allen A, Chao S et al (2014) Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796. https://doi.org/10.1111/pbi.12183
Yildiz E, Pinar H, Uzun A, Yaman M, Sumbul A, Ercisli S (2021a) Identification of genetic diversity among Juglans regia L. genotypes using molecular, morphological, and fatty acid data. Genet Res Crop Evol 68:1425–1437. https://doi.org/10.1007/s10722-020-01072-6
Yildiz A, Hajyzadeh M, Ozbek K, Nadeem MA, Hunter D (2021b) Molecular characterisation of the oldest domesticated turkish einkorn wheat landraces with simple sequence repeat (SSR) markers. Biotechnol Biotechnol Equip 35:1291–1300. https://doi.org/10.1080/13102818.2021.1970023
Zeb B, Khan I, Ali S, Bacha S, Mumtaz S, Swati Z (2009) Study on genetic diversity in pakistani wheat varieties using simple sequence repeat (SSR) markers. Afr J Biotech 8:4016–4019
Zhang D, Bai G, Zhu C, Yu J, Carver BF (2010) Genetic diversity, population structure, and linkage disequilibrium in US elite winter wheat. Plant Genome 3:117–127. https://doi.org/10.3835/plantgenome2010.03.0004
Funding
No funding was received to conduct this study.
Author information
Authors and Affiliations
Contributions
SD and FD conceptualized and established the methodology, while SD performed molecular characterization. FD analyzed the statistical data. The manuscript was written by SD and FD.
Corresponding author
Ethics declarations
Confict of interest
The author’s confirm that this article content has no confict of interest.
Consent to participate and Publish
The authors reviewed the manuscript and expressed a desire to publish this study.
Ethics approval
This article does not contain any animal or human participant investigations conducted by the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Demirel, S., Demirel, F. Molecular identification and population structure of emmer and einkorn wheat lines with different ploidy levels using SSR markers. Genet Resour Crop Evol 71, 363–372 (2024). https://doi.org/10.1007/s10722-023-01627-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01627-3