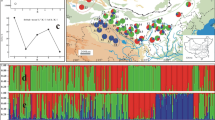
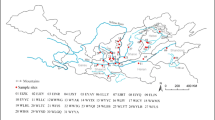
Abstract
Juglans regia is an important perennial crop cultivated for its high-quality nuts and wood. It is generally believed that J. regia survived and expanded in almost completely isolated stands in Asia after the last glaciation. Humans subsequently dispersed J. regia through cultural expansion and trade. We evaluated the spatial genetic structure and genetic diversity of 2,929 J. regia samples from 150 populations using 14 Simple Sequence Repeats (SSRs) markers. Our study revealed that regions with the highest genetic diversity included Southern Asia, Western Asia, Western Europe, and China, as illustrated using a Geostatistical Inverse Distance Weighting (IDW) interpolation of observed heterozygosity (HO), expected (HE) heterozygosity (HE), percentage of polymorphic loci (PPL), the total number of alleles (NA), and Allelic richness (RS) in Arc Geographic Information System (ArcGIS). The ecological Niche Model (ENM) showed J. regia had a high probability of association with Central Asian and Eastern Asian habitats. Population genetic structure, phylogeny, and Principal Coordinate Analysis (PCoA) identified three genetic groups corresponding to three geographic sources. Turkish and Georgian populations served as a bridge between Asian populations and Europe populations. We suggest that J. regia evolved in central Asian mountain ranges ~ 65 million years ago (Mya) and dispersed across Eurasia during climate shifts (~ 65 to 3Mya). The population contracted into multiple refugia during the Last Glacial Maximum. The current distribution of J. regia across Eurasia was shaped by the cumulative effects of contraction or expansion of different refugia and human exploitation after LGM.






Similar content being viewed by others
References
Antao T, Lopes A, Lopes RJ, Beja Pereira A, Luikart G (2008) Lositan: a workbench to detect molecular adaptation based on a FST-outlier method. BMC Bioinform 9:323
Aradhya M, Woeste K, Velasco D (2009) Genetic diversity, structure and differentiation in cultivated walnut (Juglans regia L.). In, VI Int Walnut Symp 861:127–132
Aradhya M, VelascoD., Ibrahimov Z, Toktoraliev B, Maghradze D, Musayev, M., Bobokashvili, Z., Preece, J.E. 2017. Genetic and ecological insights into glacial refugia of walnut (Juglans regia L.). Plos One 12, e0185974
Bai WN, Liao WJ, Zhang DY (2010) Nuclear and chloroplast DNA phylogeography reveal two refuge areas with asymmetrical gene flow in a temperate walnut tree from east Asia. New Phytol 188:892–901
Beer R, Kaiser F, Schmidt K, Ammann B, Carraro G, Grisa E, Tinner W (2008) Vegetation history of the walnut forests in Kyrgyzstan (Central Asia): natural or anthropogenic origin? Quat Sci Rev 27:621–632
Beerli P (2005) Comparison of bayesian and maximum likelihood inference of population genetic parameters. Bioinformatics 22:341–345
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. P Natl Acad Sci 98:4563–4568
Bernard A, Barreneche T, Lheureux F, Dirlewanger E (2018) Analysis of genetic diversity and structure in a worldwide walnut (Juglans regia L.) germplasm using SSR markers. PLoS ONE 13:e0208021
Chen L, Ma Q, Chen Y, Wang B, Pei D (2014) Identification of major walnut cultivars grown in China based on nut phenotypes and SSR markers. Sci Hortic 168:240–248
Cheviron ZA, Brumfield RT (2009) Migration selection balance and local adaptation of mitochondrial haplotypes in rufous collared sparrows (Zonotrichia capensis) along an elevational gradient. Evolution. Int J Org Evol 63:1593–1605
Cornille A, Giraud T, Bellard C, Tellier A, Le Cam B, Smulders M, Kleinschmit J, Roldan Ruiz I, Gladieux P (2013) Postglacial recolonization history of the European crabapple (Malus sylvestris M ill.), a wild contributor to the domesticated apple. Mol Ecol 22:2249–2263
Dang M, Zhang T, Hu Y, Zhou H, Woeste KE, Zhao P (2016) De novo assembly and characterization of bud, leaf and flowers transcriptome from Juglans regia L. for the identification and characterization of new EST-SSRs. Forests 7(10):247
Dangl GS, Woeste K, Aradhya MK, Koehmstedt A, Simon C, Potter D, Leslie CA, McGranahan G (2005) Characterization of 14 microsatellite markers for genetic analysis and cultivar identification of walnut. J Am Soc Hortic Sci 130:348–354
Doyle J, Doyle J (1987) Genomic plant DNA preparation from fresh tissue CTAB method. Phytochem Bull 19:11–15
Earl DA (2012) Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ebrahimi A, Zarei A, LawsonS., Woeste K, Smulders M (2016) Genetic diversity and genetic structure of Persian walnut (Juglans regia) accessions from 14 European, African, and Asian countries using SSR markers. Tree Genet. Genomes 12, 114. In
Elith J, Graham H, Anderson C, Dudík P, Ferrier R, Guisan M, Hijmans SA, Huettmann FR, Leathwick J, Lehmann A (2006) Novel methods improve prediction of species distributions from occurrence data. Ecography 29:129–151
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Feng X, Zhou H, Zulfiqar S, Luo X, Hu Y, Feng LI, Malvolti ME, Woeste K, Zhao P (2018) The phytogeographic history of common walnut in China. Front Plant Sci 21:1399
Freamo H, Oreilly P, Berg PR, Lien S, Boulding EG (2011) Outlier SNPs show more genetic structure between two bay of fundy metapopulations of Atlantic salmon than do neutral SNPs. Mol Ecol Resour 11:254–267
Fréville H, Justy F, Olivieri I (2001) Comparative allozyme and microsatellite population structure in a narrow endemic plant species, Centaurea corymbosa Pourret (Asteraceae). Mol Ecol 10:879–889
Fyfe RM, De Beaulieu JL, Binney H, Bradshaw RH, Brewer S, Le Flao A, Finsinger W, Gaillard MJ, Giesecke T, Romera G (2009) The European pollen database: past efforts and current activities. Veg Hist Archaeobot 18:417–424
Gobert S (2012) Fresh water ostracods as paleoenvironmental proxies in the Moervaart depression, a palaeo lake in sandy flanders (NW Belgium). Thesis submitted to the University of Ghent, Belgium
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Gunn BF, Aradhya M, Salick JM, Miller AJ, Yongping Y, Lin L, Xian H (2010) Genetic variation in walnuts (Juglans regia and J. sigillata; Juglandaceae): species distinctions, human impacts, and the conservation of agrobiodiversity in Yunnan. China Am J Bot 97:660–671
Han H, Woeste KE, Hu Y, Dang M, Zhang T, Gao XX, Zhou H, Feng X, Zhao G, Zhao P (2016) Genetic diversity and population structure of common walnut (Juglans regia) in China based on EST-SSRs and the nuclear gene phenylalanine ammonia lyase (PAL). Tree Genet Genomes 12:111
Helyar SJ, Hemmer HJ, Bekkevold D, Taylor M, Ogden R, Limborg M, Cariani A, Maes G, Diopere E, Carvalho G (2011) Application of SNPs for population genetics of nonmodel organisms: new opportunities and challenges. Mol Ecol Resour 11:123–136
Hemery GE, Popov SI (1998) The walnut (Juglans regia L.) forests of Kyrgyzstan and their importance as a genetic resource. Commonw for Rev 251:272–276
Hemery G, Savill P, Thakur A (2005) Height growth and flushing in common walnut (Juglans regia L.): 5 year results from provenance trials in Great Britain. Forestry 78:121–133
Hengl T (2009) A practical guide to geostatistical mapping. Hengl Amsterdam
Holland MM, Parson W (2011) GeneMarker® HID: a reliable software tool for the analysis of forensic STR data. J Forensic Sci 56:29–35
Kalinowski ST (2005) hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4:981
Maguire T, Peakall R, Saenger P (2002) Comparative analysis of genetic diversity in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae) detected by AFLPs and SSRs. Theor Appl Genet 104:388–398
Malvolti M, Taurchini D, Fineschi S, Beritognolo I, Maccaglia E, Cannata F, Fornari B (1999) Isozyme and organellar DNA analysis of genetic diversity in natural/naturalised European and Asiatic walnut (Juglans regia L.) populations. In, IV Int Walnut Symp 544:167–178
Manel S, Holderegger R (2013) Ten years of landscape genetics. Trends Ecol Evol 28:614–621
Manni F, Guerard E, Heyer E (2004) Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier’s algorithm. Hum Biol 76:173–190
McGranahan G, Leslie C (1991) Walnuts (Juglans). Genet Resour Temp Fruit Nut Crops 290:907–974
Miller HC, Allendorf F, Daugherty CH (2010) Genetic diversity and differentiation at MHC genes in island populations of tuatara (Sphenodon spp.). Mol Ecol 19:3894–3908
Nei M (1973) Analysis of gene diversity in subdivided populations. P Natl Acad Sci 70:3321–3323
Ornelas JF, Gándara E, Vásquez Aguilar AA, Ramírez Barahona S, Ortiz Rodriguez AE, González C, Saules MTM, Ruiz SE (2016) A mistletoe tale: postglacial invasion of Psittacanthus chiedeanus (Loranthaceae) to Mesoamerican cloud forests revealed by molecular data and species distribution modeling. BMC Evol Biol 16:78
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Phillips SJ, Dudík M (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31:161–175
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Pollegioni P, Woeste K, Major A, Scarascia Mugnozza G, Malvolti ME (2009) Characterization of Juglans nigra (L.), Juglans regia (L.) and Juglans x intermedia (Carr.) by SSR markers: a case study in Italy. Silvae Genet 58:68–78
Pollegioni P, Woeste K, Olimpieri I, Marandola D, Cannata F, Malvolti ME (2011) Long term human impacts on genetic structure of Italian walnut inferred by SSR markers. Tree Genet Genomes 7:707–723
Pollegioni P, Woeste KE, Chiocchini F, Olimpieri I, Tortolano V, Clark J, Hemery GE, Mapelli S, Malvolti ME (2014) Landscape genetics of Persian walnut (Juglans regia L.) across its Asian range. Tree Genet Genomes 10:1027–1043
Pollegioni P, Woeste KE, Chiocchini F, Del Lungo S, Olimpieri I, Tortolano V, Clark J, Hemery GE, Mapelli S, Malvolti ME (2015) Ancient humans influenced the current spatial genetic structure of common walnut populations in Asia. PLoS ONE 10:e0135980
Pollegioni P, Woeste K, Chiocchini F, Del Lungo S, Ciolfi M, Olimpieri I, Tortolano V, Clark J, Hemery GE, Mapelli S (2017) Rethinking the history of common walnut (Juglans regia L.) in Europe: Its origins and human interactions. PLoS ONE 12:e0172541
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rink G, Carroll ER, Kung FH (1989) Estimation of Juglans nigra L. mating system parameters. For Sci 35:623–627
Roor W, Konrad H, Mamadjanov D, Geburek T (2017) Population differentiation in common walnut (Juglans regia L.) across major parts of its native range insights from molecular and morphometric data. J Hered 108:391–404
RosenbergN A (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Savolainen O, Pyhäjärvi T, Knürr T (2007) Gene flow and local adaptation in trees. Annu Rev Ecol Evol Syst 38:595–619
Slatkin M, Barton NH (1989) A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43:1349–1368
Takezaki N, Nei M, Tamura K (2009) POPTREE2: software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol Biol Evol 27:747–752
Team RC (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna http .www.R-project.org
Torokeldiev N, Ziehe M, Gailing O, Finkeldey R (2019) Genetic diversity and structure of natural Juglans regia L. populations in the southern Kyrgyz Republic revealed by nuclear SSR and EST-SSR markers. Tree Genet Genomes 15:1–12
Tsuda Y, Nakao K, Ide Y, Tsumura Y (2015) The population demography of B. etula maximowicziana, a cool temperate tree species in Japan, in relation to the last glacial period: its admixture like genetic structure is the result of simple population splitting not admixing. Mol Ecol 24:1403–1418
Turvey ST, Tong H, Stuart AJ, Lister AM (2013) Holocene survival of late pleistocene megafauna in China: a critical review of the evidence. Quat Sci Rev 76:156–166
Vahdati K (2013) Traditions and folks for walnut growing around the silk road. In: international symposium on fruit sulture and its traditional knowledge along silk road countries 1032: 19-24
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wang WT, Xu B, Zhang DY, Bai WN (2016) Phylogeography of postglacial range expansion in Juglans mandshurica (Juglandaceae) reveals no evidence of bottleneck, loss of genetic diversity, or isolation by distance in the leading edge populations. Mol Phylogenet Evol 102:255–264
Waples RS, Gaggiotti O (2006) INVITED REVIEW: what is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15:1419–1439
Warren DL, Glor RE, Turelli M (2008) Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolut Int J Org Evolut 62:2868–2883
Warren DL, Glor RE, Turelli M (2010) ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33:607–611
Weckerle C, Huber FK, Yang Y, Song W (2005) Walnuts among the Shuhi in Shuiluo, eastern Himalayas. Econ Bot 59:287–290
Woeste K, Burns R, Rhodes O, Michler C (2002) Thirty polymorphic nuclear microsatellite loci from black walnut. J Hered 93:58–60
Woeste K, Michler C (2011) Juglans. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, forest trees. Springer, Berlin
Yuan XY, Sun YW, Bai XR, Dang M, Feng XJ, Zulfiqar S, Zhao P (2018) Population structure, genetic diversity, and gene introgression of two closely related walnuts (Juglans regia and J. sigillata) in southwestern China revealed by EST-SSR markers. Forests 9:646
Zhao P, Woeste KE (2011) DNA markers identify hybrids between butternut (Juglans cinerea L.) and Japanese walnut (Juglans ailantifolia Carr.). Tree Genet Genomes 7:511–533
Zohary D, Hopf M, Weiss E (2012) Domestication of Plants in the old world: the origin and spread of domesticated plants in Southwest Asia, Europe, and the mediterranean Basin. Oxford University Press on Demand, Oxford
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP-2023R369), King Saud University, Riyadh, Saudi Arabia.
Funding
This work was supported by the National Natural Science Foundation of China (No. 41471038; No. 31200500), the Program for Excellent Young Academic Backbones funding by Northwest University, Shaanxi Academy of Science Research Funding Project (2019 K-06), and Natural Science Foundation of Shaanxi Province of China (2019JM-008). We thank N. Hou, Y. L. Xu, N. Lin, and L. Wang for assisting with sampling. Mention of a trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture and does not imply its approval to the exclusion of other products or vendors that also may be suitable.
Author information
Authors and Affiliations
Contributions
PZ conceived the idea and conducted research. The investigation was carried out by HK and IU. Data analysis was carried out by HK, KW, ME. M, MY. K A. A, UZ, and SF provided technical expertise. AA, SS, IU, MY, and KW helped in the writing of the original draft. AI. G, A A. M and FC helped in literature improvement, data re-analysis, language modification, funding acquisition and revision of manuscript. All authors carefully read, revise, and approved the article for submission.
Corresponding authors
Ethics declarations
Conflict of interest
There is no conflict of interest and all the authors have significantly contributed to the overall manuscript preparation.
Ethics approval
Experimental research and field studies on plants (either cultivated or wild), including the collection of plant material, complied with relevant institutional, national, and international guidelines and legislation. We also took appropriate permission from the relevant research section of the University during the specimens’ collection and experimentation. We confirm that during the collection and execution of the experiment, the authors have complied with the IUCN Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khan, H., Ullah, I., Woeste, K. et al. Population genetics informs new insights into the phytogeographic history of Juglans regia L.. Genet Resour Crop Evol 70, 2263–2278 (2023). https://doi.org/10.1007/s10722-023-01597-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01597-6