Abstract
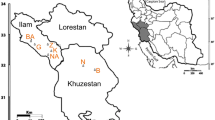
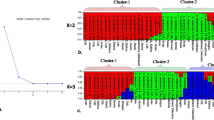
The genetic diversity of 34 A. desertorum accessions was studied using 28 ISSR primers. A total of 448 loci were amplified in the A. desertorum genome, and 402 (90%) of them were polymorphic. The primer (AG)8YC produced the greatest number of polymorphic fragments, while the primer (AC)8YA produced the lowest number. The number of effective alleles ranged between 1.26 and 1.57. The highest value of Shanon index belonged to primer (GA)8T, while the highest value of Nei index (0.33) was detected for primers (GA)8T, (TC)8C, and (GACA)4. Primer (GA)8GCC with PIC = 0.40 was the best informative marker in the evaluation of A. desertorum genetic diversity. Genetic similarity among studied accessions was between 0.45 (G03 and G17) and 0.80 (G01 and G11). Using Bayesian and WPGMA classification algorithms, the A. desertorum germplasm was divided into two major subgroups (Red and Green), which consisted of 13 and 17 accessions, respectively, and also admixture accessions. Late-maturing accessions were distinguished from early-maturing accessions and mostly localized in the Green subgroup. The Red subgroup had more private alleles as well as heterozygosity compared with the Green subgroup and therefore also had promising selection potential. Totally, ISSR markers could distinguish early mature genotypes, and this is significant for increased forage yield. Likewise, the recognized heterotic groups could help breeders select highly polymorphic genotypes in polycross breeding.


Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Absattar T, Absattarova A, Fillipova N, Otemissova A, Shavrukov Y (2018) Diversity array technology (DArT) 56K analysis, confirmed by SNP markers, distinguishes one crested wheatgrass Agropyron species from two others found in Kazakhstan. Mol Breeding 38:37
Alam MA, Juraimi AS, Rafii MY, Hamid AA, Arolu IW, Abdul Latif M (2015) Genetic diversity analysis among collected purslane (Portulaca oleracea L.) accessions using ISSR markers. CR Biol 338:1–11
Arghavani A, Asghari A, Shokrpour M, Mohammaddost C (2010) Genetic diversity in ecotypes of two Agropyron species using RAPD markers. Res J Environ Sci 4:50–56
Ayaz A, Zaman W, Saqib S, Ullah F, Mahmood T (2020) Phylogeny and diversity of lamiaceae based on rps14 gene in Pakistan. Genetika 52:435–452
Bor NL (1970) Gramineae. In: Iranica Flora, Rechinger KH (eds) Akademische Druk-u. Verlagsanstalt, Graz, pp 571–573
Che Y, Yang Y, Yang X, Li X, Li L (2015) Phylogenetic relationship and diversity among Agropyron Gaertn. germplasm using SSRs markers. Plant Syst Evol 301:163–170
Copete A, Moreno R, Cabrera A (2018) Characterization of a world collection of Agropyron cristatum accessions. Genet Resour Crop Evol 65:1455–1469
Doyle J, Doyle J (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Farshadfar M, Nowrozi R, Safari H, Shirvani H, Amjadian M (2018) Assessment of genetic diversity in Agropyron desertorum accessions using ISSR molecular marker. Futur of Food J Food Agric Soc 6:20–29
Frankham R, Briscoe DA, Ballou JD (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Godwin ID, Aitken EA, Smith LW (1997) Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis 18:1524–8
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int 2015:431487
Han H, Qiu R, Liu Y, Zhou X, Gao C, Pang Y, Zhao Y (2022) Analysis of chloroplast genomes provides insights into the evolution of Agropyron. Front Genet 13:832809
Hayward AC, Tollenaere R, Dalton-Morgan J, Batley J (2015) Molecular marker applications in plants. Methods Mol Biol 1245:13–27
Jafari AA, Seydemohammadi AR, Abdi N, Madah AH (2008) Seed and hay production in 31 genotypes of desert wheatgrass (Agropyron desertorum) using drought tolerance indices. Iran J Range Desert Res 15:114–128
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Li H, Jiang B, Wang J, Lu Y, Zhang J, Pan C, Yang X, Li X, Liu W, Li L (2017) Appling of novel powdery mildew resistance genes from Agropyron cristatum chromosome 2P’. Theor Appl Genet 130:109–121
Mohammadi R, Panahi B, Amiri S (2020) ISSR Based study of fine fescue (Festuca ovina L.) highlighted the genetic diversity of Iranian accessions. Cytol Genet 54:257–263
Mohammadi R, Amiri S, Montakhabi Kalajahi V (2022) ISSR markers efficiency to assess cool-season grass species genetic diversity and phylogenetic relationships. Proc Natl Acad Sci India Sect B Biol Sci 92:691–699
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Novembre J (2016) Pritchard, Stephens, and Donnelly on population structure. Genetics 204:391–393
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. Accessed from. https://darwin.cirad.fr/
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rahnemoun B, Hatami Maleki H, Mohammadi R (2018) Genetic variability in different accessions of Agropyron based on morphological traits. Modares J Biotechnol 9:517–523
Razi M, Amiri ME, Darvishzadeh R, Doulati Baneh H, Alipour H, Martínez-Gómez P (2020) Assessment of genetic diversity of cultivated and wild Iranian grape germplasm using retrotransposon-microsatellite amplified polymorphism (REMAP) markers and pomological traits. Mol Biol Rep 47:7593–7606
Reddy MP, Sarla N, Siddiq EA (2002) Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 128:9–17
Sakamoto S (1964) Cytogenetical problems in Agropyron hybrids. Seiken Jiho 16:38–47
Salgotra RK, Stewart CN (2020) Functional markers for precision plant breeding. Int J Mol Sci 21:4792
Serrote CML, Reiniger LRS, Silva KB, Rabaiolli SMDS, Stefanel CM (2019) Determining the polymorphism information content of a molecular marker. Gene 5:144175
Spataro G, Tiranti B, Arcaleni P, Bellucci E, Attene G, Papa R, Zeuli PS, Negri V (2011) Genetic diversity and structure of a worldwide collection of Phaseolus coccineus L. Theor Appl Genet 122:1281–1291
Wu Q, Zang F, Ma Y, Zheng Y, Zang D (2020) Analysis of genetic diversity and population structure in endangered Populus wulianensis based on 18 newly developed EST-SSR markers. Glob Ecol Conserv 24:e01329
Yeh FC, Yang RC, Boyle T (1999) POPGENE version 1.31, Microsoft Window-based freeware for population genetic analysis. University of Alberta and Centre for International Forestry Research
Acknowledgment
The authors thank the University of Maragheh and the Agricultural Biotechnology Research Institute of Iran for lab facilities.
Funding
This work was supported by university of Maragheh and Author Hamid Hatami Maleki has received research support from University of Maragheh.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by RM and HHM, MH, MA, MR and HHM. The first draft of the manuscript was written by HHM and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Maleki, H.H., Mohammadi, R., Arshad, M. et al. Genetic variability and population structure of Agropyron desertorum accessions from Iran based on inter simple sequence repeat (ISSR) assay. Genet Resour Crop Evol 70, 2511–2520 (2023). https://doi.org/10.1007/s10722-023-01579-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01579-8