Abstract
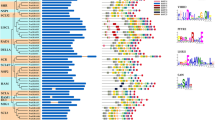
The domain of unknown function (DUF221 domain-containing) proteins regulates various aspects of plant growth, development, responses to abiotic stresses, and hormone transduction pathways. A comprehensive genome-wide analysis was performed in its genome to understand the role of DDP genes (DUF221) in the common bean (Phaseolus vulgaris L.). A total of 12 DDP genes were identified and distributed in 8 chromosomes in the common bean genome. The physical and biochemical characteristics of amino acids, motif and intron–exon structure, and cis-regulatory elements of DDP members were determined. Phylogenetically all PvDDPs were clustered into nine clades, subsequently supported by their gene structure and conserved motifs distribution. The PvDDPs contained various cis-acting elements involved in plant responses to abiotic and various phytohormones stresses. A total of 45 different cis-regulatory elements in the putative promoter regions of the PvDDPs were identified. ERE and ABRE were discovered to be present in all PvDDPs, indicating that they may be regulated by ethylene and ABA, both of which are strongly associated with biotic stress response in plant species. Additionally, PvDDPs were targeted by multiple miRNA gene families as well. In this context, the most targeted DDP family members are PvDD10 and PvDDP11. The miRNA target analysis showed that Pvu-miR2594, Pvu-miR169, Pvu-miR2584, Pvu-miR530, Pvu-miR156, and Pvu-miR2592 target these genes. There is a strong correlation between abiotic stress and PvDDPs expression in both leaf and root tissues. PvDDP11 is the unique and highest upregulated gene with hormone treatment and abiotic stress among all the members. Expression of the PvDDP11 gene indicated a strong correlation with drought and salt stress in the common bean roots and leaves, respectively. In conclusion, this study predicted that the putative DDP genes might help improve abiotic and phytohormone tolerance in common bean.









Similar content being viewed by others
Data availability
The datasets analysed during the current study are available in the NCBI/SRA. Database with following accession numbers; PRJNA508605 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA508605), PRJNA327176 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA327176), PRJNA574280 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA574280), PRJNA691982 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA691982), PRJNA656794 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA656794), PRJNA558376 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA558376), and PRJNA574280 (https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA574280).
References
Aalto MK, Helenius E, Kariola T, Pennanen V, Heino P, Hõrak H, Puzõrjova I, Kollist H, Palva ET (2012) ERD15—An attenuator of plant ABA responses and stomatal aperture. Plant Sci 182:19–28. https://doi.org/10.1016/j.plantsci.2011.08.009I
Alamillo JM, Díaz-leal JL, Sánchez-moran MV, Pineda M (2010) Molecular analysis of ureide accumulation under drought stress in Phaseolus vulgaris L. Plant Cell Environ 33:1828–1837. https://doi.org/10.1111/j.1365-3040.2010.02187.xI
Bateman A, Coggill P, Finn RD (2010) DUFs: families in search of function. Acta Cryst 66:1148–1152. https://doi.org/10.1107/S1744309110001685I
Bayuelo Jimenez J, Jasso Plata N, Ochoa I (2012) Growth and physiological responses of Phaseolus species to salinity stress. Int J Agron. https://doi.org/10.1155/2012/527673I
Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.)–model food legumes. Plant Soil 252:55–128. https://doi.org/10.1023/A:1024146710611I
Cao X, Yang KZ, Xia C, Zhang XQ, Chen LQ, Ye D (2009) Characterization of DUF724 gene family in Arabidopsisthaliana. Plant Mol Biol 72:61. https://doi.org/10.1007/s11103-009-9551-5I
Chen C, Chen H, He Y, Xia R (2018) TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv:289660. Doi: https://doi.org/10.1101/289660I
Chou KC, Shen HB (2008) Cell-PLoc: a package of web servers for predicting subcellular localization of proteins in various organisms. Nat Protoc 3:153. https://doi.org/10.1038/nprot.2007.494I
Cortés AJ (2013) On the Origin of the Common Bean (Phaseolus vulgaris L.). Am J Plant Sci 4 (10):1998–2000. https://doi.org/10.4236/ajps.2013.410248
Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46:W49–W54. https://doi.org/10.1093/nar/gky316I
Enright A, John B, Gaul U, Tuschl T, Sander C, Marks D (2003) MicroRNA targets in Drosophila. Genome Biol 4:1–27. https://doi.org/10.1186/gb-2003-4-11-p8I
Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR (2015) HMMER web server: 2015 update. Nucleic Acids Res 43:W30–W38. https://doi.org/10.1093/nar/gkv397I
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador Vegas A (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. https://doi.org/10.1093/nar/gkv1344I
Ganie SA, Pani DR, Mondal TK (2017) Genome-wide analysis of DUF221 domain-containing gene family in Oryza species and identification of its salinity stress-responsive members in rice. PLoS ONE 12:e0182469. https://doi.org/10.1371/journal.pone.0182469I
Gholizadeh A (2020) Pectin methylesterase activity of plant DUF538 protein superfamily. Physiol Mol Biol Plants 26:829–839. https://doi.org/10.1007/s12298-020-00763-9I
Guo Z, Kuang Z, Wang Y, Zhao Y, Tao Y, Cheng C, Yang J, Lu X, Hao C, Wang T (2020) PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res 48:D1114–D1121. https://doi.org/10.1093/nar/gkz894I
Hesberg C, Hänsch R, Mendel RR, Bittner F (2004) Tandem orientation of duplicated xanthine dehydrogenase genes from Arabidopsis thaliana: differential gene expression and enzyme activities. J Biol Chem 279:13547–13554. https://doi.org/10.1074/jbc.M312929200I
Hiz MC, Canher B, Niron H, Turet M (2014) Transcriptome analysis of salt tolerant common bean (Phaseolus vulgaris L.) under saline conditions. PLoS ONE 9:e92598. https://doi.org/10.1371/journal.pone.0092598I
Hon J, Marusiak M, Martinek T, Kunka A, Zendulka J, Bednar D, Damborsky J (2021) SoluProt: prediction of soluble protein expression in Escherichia coli. Bioinform 37:23–28. https://doi.org/10.1093/bioinformatics/btaa1102I
Hou C, Tian W, Kleist T, He K, Garcia V, Bai F, Hao Y, Luan S, Li L (2014) DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res 24:632–635. https://doi.org/10.1038/cr.2014.14I
Jamil A, Riaz S, Ashraf M, Foolad MR (2011) Gene expression profiling of plants under salt stress. Crit Rev Plant Sci 30:435–458. https://doi.org/10.1080/07352689.2011.605739I
Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE (2005) The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res 15:1292–1297. https://doi.org/10.1101/gr.4064205I
Kariola T, Brader G, Helenius E, Li J, Heino P, Palva ET (2006) EARLY RESPONSIVE TO DEHYDRATION 15, a negative regulator of abscisic acid responses in arabidopsis. Plant Physiol 142:1559–1573. https://doi.org/10.1104/pp.106.086223I
Kavas M, Baloğlu MC, Atabay ES, Ziplar UT, Daşgan HY, Ünver T (2016) Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol Genet Genom 291:129–143. https://doi.org/10.1007/s00438-015-1095-6I
Kavas M, Kızıldoğan AK, Abanoz B (2017) Comparative genome-wide phylogenetic and expression analysis of SBP genes from potato (Solanum tuberosum). Comput Biol Chem 67:131–140. https://doi.org/10.1016/j.compbiolchem.2017.01.001I
Kavas M, Yildirim K, Secgin Z, Abdulla MF, Gokdemir G (2021) Genome-wide identification of the BURP domain-containing genes in Phaseolus vulgaris. Physiol Mol Biol Plants 27:1885–1902. https://doi.org/10.1007/s12298-021-01052-9I
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protoc 10:845–858. https://doi.org/10.1038/nprot.2015.053I
Krogh A, Larsson B, Von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol 305:567–580. https://doi.org/10.1006/jmbi.2000.4315I
Leng ZX, Liu Y, Chen ZY, Guo J, Chen J, Zhou YB, Chen M, Ma YZ, Xu ZS, Cui XY (2021) Genome-wide analysis of the DUF4228 family in soybean and functional identification of GmDUF4228 -70 in response to drought and Salt stresses. Front Plant Sci 12:628299–628299. https://doi.org/10.3389/fpls.2021.628299I
Li J, Hu E, Chen X, Xu J, Lan H, Li C, Hu Y, Lu Y (2016) Evolution of DUF1313 family members across plant species and their association with maize photoperiod sensitivity. Genom 107:199–207. https://doi.org/10.1016/j.ygeno.2016.01.003I
Li L, Xie C, Ye T, Xu J, Chen R, Gao X, Zhu J, Xu Z (2017) Molecular characterization, expression pattern and function analysis of the rice OsDUF866 family. Biotechnol Biotechnol Equip 31:243–249. https://doi.org/10.1080/13102818.2016.1268932I
Li LH, Lv MM, Li X, Ye TZ, He X, Rong SH, Dong YL, Guan Y, Gao XL, Zhu JQ, Xu ZJ (2018) The rice OsDUF810 family: OsDUF810.7 may be involved in the tolerance to salt and drought. Mol Biol 52:489–496. https://doi.org/10.1134/S002689331804012XI
Li L, Ye T, Guan Y, Lv M, Xie C, Xu J, Gao X, Zhu J, Cai L, Xu Z (2018) Genome-wide identification and analyses of the rice OsDUF936 family. Biotechnol Biotechnol Equip 32:309–315. https://doi.org/10.1080/13102818.2017.1413421I
Liu C, Xie T, Chen C, Luan A, Long J, Li C, Ding Y, He Y (2017) Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom 18:1–16. https://doi.org/10.1186/s12864-017-3896-yI
Maher C, Stein L, Ware D (2006) Evolution of arabidopsis microRNA families through duplication events. Genome Res 16:510–519. https://doi.org/10.1101/gr.4680506I
Marciniak K, Przedniczek K (2019) Comprehensive insight into gibberellin- and jasmonate-mediated stamen development. Genes 10:811. https://doi.org/10.3390/genes10100811I
Montalbini P (1995) Effect of rust infection on purine catabolism enzyme levels in wheat leaves. Physiol Mol Plant Pathol 46:275–292. https://doi.org/10.1006/pmpp.1995.1022I
Nabi RBS, Tayade R, Imran QM, Hussain A, Shahid M, Yun B-W (2021) Functional insight of nitric-oxide induced DUF genes in arabidopsis. Nitric Oxide Plants. https://doi.org/10.3389/fpls.2020.01041I
Nietzsche M, Schießl I, Börnke F (2014) The complex becomes more complex: protein-protein interactions of SnRK1 with DUF581 family proteins provide a framework for cell- and stimulus type-specific SnRK1 signaling in plants. Front Plant Sci. https://doi.org/10.3389/fpls.2014.00054I
Niron H, Barlas N, Salih B, Türet M (2020) Comparative transcriptome, metabolome, and ionome analysis of two contrasting common bean genotypes in saline conditions. Front Plant Sci. https://doi.org/10.3389/fpls.2020.599501I
Olvera Carrillo Y, Campos F, Reyes JL, Garciarrubio A, Covarrubias AA (2010) Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in arabidopsis. Plant Physiol 154:373–390. https://doi.org/10.1104/pp.110.158964I
Qiao X, Li Q, Yin H, Qi K, Li L, Wang R, Zhang S, Paterson AH (2019) Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol 20:1–23. https://doi.org/10.1186/s13059-019-1650-2I
Qiu L, Chen R, Fan Y, Huang X, Luo H, Xiong F, Liu J, Zhang R, Lei J, Zhou H, Wu J, Li Y (2019) Integrated mRNA and small RNA sequencing reveals microRNA regulatory network associated with internode elongation in sugarcane (Saccharum officinarum L.). BMC Genom 20:817. https://doi.org/10.1186/s12864-019-6201-4I
Rai A, Suprasanna P, D’Souza SF, Kumar V (2012) Membrane topology and predicted RNA-binding function of the ‘early responsive to dehydration (ERD4)’plant protein. PLoS ONE. https://doi.org/10.1371/journal.pone.0032658I
Sagi M, Omarov RT, Lips SH (1998) The Mo-hydroxylases xanthine dehydrogenase and aldehyde oxidase in ryegrass as affected by nitrogen and salinity. Plant Sci 135:125–135. https://doi.org/10.1016/S0168-9452(98)00075-2I
Shaik R, Ramakrishna W (2012) Bioinformatic analysis of epigenetic and microRNA mediated regulation of drought responsive genes in rice. PLoS ONE 7:e49331. https://doi.org/10.1371/journal.pone.0049331I
Shao H, Chen S, Zhang K, Cao Q, Zhou H, Ma Q, He B, Yuan X, Wang Y, Chen Y (2014) Isolation and expression studies of the ERD15 gene involved in drought-stressed responses. Genet Mol Res 13:10852–10862. https://doi.org/10.4238/2014.December.19.6I
Vaattovaara A, Brandt B, Rajaraman S, Safronov O, Veidenberg A, Luklová M, Kangasjärvi J, Löytynoja A, Hothorn M, Salojärvi J, Wrzaczek M (2019) Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun Biol. https://doi.org/10.1038/s42003-019-0306-9I
Wang L, Shen R, Chen LT, Liu YG (2014) Characterization of a novel DUF1618 gene family in rice. J Integr Plant Biol 56:151–158. https://doi.org/10.1111/jipb.12130I
Wang T, Chen L, Zhao M, Tian Q, Zhang W-H (2011) Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom 12:367. https://doi.org/10.1186/1471-2164-12-367I
Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-h, Jin H, Marler B, Guo H (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49–e49. https://doi.org/10.1093/nar/gkr1293I
Waseem M, Aslam MM, Shaheen I (2021) The DUF221 domain-containing (DDP) genes identification and expression analysis in tomato under abiotic and phytohormone stress. GM Crop Food. https://doi.org/10.1080/21645698.2021.1962207I
Watanabe S, Matsumoto M, Hakomori Y, Takagi H, Shimada H, Sakamoto A (2014) The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ 37:1022–1036. https://doi.org/10.1111/pce.12218I
Yang Q, Niu X, Tian X, Zhang X, Cong J, Wang R, Zhang G, Li G (2020) Comprehensive genomic analysis of the DUF4228 gene family in land plants and expression profiling of ATDUF4228 under abiotic stresses. BMC Genom 21:12. https://doi.org/10.1186/s12864-019-6389-3I
Yuan F, Yang H, Xue Y, Kong D, Ye R, Li C, Zhang J, Theprungsirikul L, Shrift T, Krichilsky B (2015) OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature, vol 514, pg 367
Zhong H, Zhang H, Guo R, Wang Q, Huang X, Liao J, Li Y, Huang Y, Wang Z (2019) Characterization and functional divergence of a novel DUF668 gene family in rice based on comprehensive expression patterns. Gene 10:980. https://doi.org/10.3390/genes10120980I
Zhou X, Zhu X, Shao W, Song J, Jiang W, He Y, Yin J, Ma D, Qiao Y (2020) Genome-wide mining of wheat DUF966 gene family provides new insights into salt stress responses. Front Plant Sci. https://doi.org/10.3389/fpls.2020.569838I
Funding
This study was supported by the Research Fund of Ondokuz Mayıs University (PYO.ZRT.1901.17.010).
Author information
Authors and Affiliations
Contributions
MK planned, designed, and performed the experiments, KA, ZS, BAY and GG analyzed the data, wrote the paper; KY wrote the paper. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests. The funding sponsors had no role in the design of the study, collection, analyses, interpretation of the data, writing of the manuscript, and in the decision to publish the results.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

10722_2022_1421_MOESM1_ESM.jpg
Supplementary file1 (JPG 19070 kb): Fig. S1 Multiple alignments of amino acid sequences of DDPs from P. vulgaris. Among the members, many highly conserved amino acid sites were identified
10722_2022_1421_MOESM2_ESM.png
Supplementary file2 (PNG 1971 kb): Fig. S2 3D structure of PvDDP proteins visualized using Phyre2 server with an intensive mode
10722_2022_1421_MOESM6_ESM.xlsx
Supplementary file6 (XLSX 10 kb): Table S3 One-to-one orthologous relationships between common bean and Arabidopsis as well as that between common bean and rice
10722_2022_1421_MOESM8_ESM.xlsx
Supplementary file8 (XLSX 13 kb): Table S5 Predicted functional partners of PvDDPs and functional enrichments in the network
Rights and permissions
About this article
Cite this article
Kavas, M., Mostafa, K., Seçgin, Z. et al. Genome-wide analysis of DUF221 domain-containing gene family in common bean and identification of its role on abiotic and phytohormone stress response. Genet Resour Crop Evol 70, 169–188 (2023). https://doi.org/10.1007/s10722-022-01421-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01421-7