Abstract
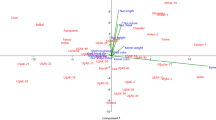
In this study, walnut genotypes that were selected during two growing seasons among thousands of seedlings were analyzed in terms of detailed morphometric, phenological, and chemical traits. A multivariate analysis was conducted with valuable traits for breeding and selection such as morphometric traits, chemical composition, and phenological characteristics. Also, genotypes were characterized by a retrotransposon-based iPBS marker system. The correlation analysis showed significant positive and negative correlations between agro-morphological characters. The principal component analysis explained 71.44% of the total variance into five main components. Principal component and hierarchical cluster analysis divided genotypes into three groups and identified subgroups based on both agro-morphological characters and iPBS marker systems. A high level of polymorphism ratio was observed for tested markers. Mantel’s test demonstrated relatively low correlations between molecular and morphological treats (r = 0.04). The genetic similarities among all individuals ranged from 0.39 (between 018 and 015 or 045 genotypes) to 0.98 (between 090 and 094 genotypes) with a mean similarity of 0.67. Remarkable phenotypic and molecular variations were observed among the genotypes. The features of some investigated genotypes were above the acceptable thresholds for walnut selection in breeding programs, and our study indicated that iPBS markers can be beneficial in walnut breeding programs, allowing the evaluation of the genetic relationship between genotypes, helping to differentiate and select the best genotypes to improve agronomic properties.






Similar content being viewed by others
References
Akça Y (2016) Walnut production. Anıt Matbaası
Allaire J (2012) R Studio: integrated development environment for R. Boston, MA 770(394):165–171
Amiri R, Vahdati K, Monseripoor S, Mozaffari MR, Leslie C (2010) Correlations between some horticultural traits in walnut. HortScience 45:1690–1694
AOAC (1990) Official Methods of Analysis, 15th edn. AOAC, Washsington, DC
AOAC (1995) Official methods of analysis, 16th edn. Association of Offical Analytical Chemists, Washington DC
Aydın F, Özer G, Alkan M, Çakır İ (2020) The utility of iPBS retrotransposons markers to analyze genetic variation in yeast. Int J Food Microbiol 325:108647. https://doi.org/10.1016/j.ijfoodmicro.2020.108647
Badenes ML, Parfitt DE (1998) Phylogeny of the genus pistacia as determined from analysis of the chloroplast genome. Proc Natl Acad Sci USA 7:25–26
Boronnikova SV, Kalendar RN (2010) Using IRAP markers for analysis of genetic variability in populations of resource and rare species of plants. Russ J Genet 46:36–42
Castro I, D’Onofrio C, Martín JP, Marı´a Ortiz J, De Lorenzis G, Ferreira V, Pinto-Carnide O (2012) Effectiveness of AFLPs and retrotransposon-based markers for the Identification of portuguese grapevine cultivars and clones. Mol Biotechnol 52:26–39
Christopoulos MV, Rouskas D, Tsantili E, Bebeli PJ (2010) Germplasm diversity and genetic relationships among walnut (Juglans regia L.) cultivars and Greek local selections revealed by Inter Simple Sequence Repeat (ISSR) marker. Sci Hortic 125(4):584–592
Cosmulescu S, Botu M (2012) Walnut biodiversity in South-Western Romania-resource for perspective cultivars. Pak J Bot 44(1):307–311
Doğan Y, Kafkas S, Sütyemez M, Akça Y, Türemiş N (2014) Assessment and characterization of genetic relationships of walnut (Juglans regia L.) genotypes by three types of molecular markers. Sci Hortic 168:81–87
El Zayat MAS, Hassan AH, Nishawy E, Ali M, Amar MH (2021) Patterns of genetic structure and evidence of Egyptian citrus rootstock based on informative SSR, LTR-IRAP and LTR-REMAP molecular markers. J Genet Eng Biotechnol 19(1):29
Erper I, Ozer G, Kalendar R, Avci S, Yildirim E, Alkan M, Turkkan M (2021) Genetic diversity and pathogenicity of Rhizoctonia spp. isolates associated with red cabbage in Samsun (Turkey). J Fungi 7(3):234
Farrokhi Toolir J, Mozaffari M (2020) Morphological study of some persian walnut genotypes and commercial cultivars cultured in Kerman region in southeast of Iran. Agric Conspec Sci 85(2):123–137
Finnegan DJ (1989) Eukaryotic transposable elements and genome evolution. Trends Genet 5:103–107
Fjellstrom RG, Parfitt DE, McGranahan GH (1994) Genetic relationships and characterization of Persian walnut (J. regia L.) cultivars using restriction fragment length polymorphisms (RFLPs). J Am Soc Hortic Sci 119(4):833–839
Hashemi SM, Khadivi A (2020) Morphological variability of Prunus lycioides Spach germplasm using multivariate analysis. Sci Hort 261:108973
Houmanata K, Abdellaha K, Hssaini L, Razouk R, Hanine H, Jaafarya S, Charafi J (2021) Molecular diversity of walnut (Juglans regia L.) among two major areas in Morocco in contrast with foreign varieties. Int J Fruit Sci 21(1):180–192
İpek M, Arıkan Ş, Pırlak M, Eşitken A (2019) Phenological, morphological and molecular charecterization of some promising walnut (Juglans regia L.) genotypes in Konya. Erwerbs-Obstbau 61:149–156
Kabiri G, Bouda S, Elhansali M, Haddioui A (2019) Biochemical characterization and antioxidant activity of walnut kernel (Juglans regia L.) of accessions from Middle and High Atlas in Morocco. Acta Sci Biol Sci 41:1–12
Kafkas S, Özkan H, Sütyemez M (2005) DNA polymorphism and assessment of genetic relationships in walnut genotypes based on AFLP and SAMPL markers. J Am Soc Hortic Sci 130(4):585–590
Kalendar R, Schulman A (2006) IRAP and REMAP for retrotranspozon-based genotyping and fingerprinting. Nat Protoc 1:2478–2484
Kalendar R, Antonius K, Smýkal P, Schulman AH (2010) iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor Appl Genet 121(8):1419–1430
Karadağ H, Akça Y (2011) Phenological and pomological properties of promising walnut (Juglans regia L.) genotypes from selected native population in Amasya Province. Afr J Biotechnol 10(74):16763–16768
Kaya E, Yılmaz-Gokdoğan E (2016) Using two retrotransposon based marker systems (IRAP and REMAP) for molecular characterization of olive (Olea europaea L.) cultivars. Not Bot Horti Agrobot 44(1):167–174
Khadivi A, Montazerana A, Rezaeib M, Ebrahimic A (2019) The pomological characterization of walnut (Juglans regia L.) to select the superior genotypes – An opportunity for genetic improvement. Sci Hortic 248:29–33
Kırdök E, Çiftçi YÖ (2016) Retrotransposon marker systems as a tool to analyze molecular diversity of mediterranean Pistacia species. J Agric Biol 18:601–606
Kumar LS (1999) DNA markers in plant improvement: an overview. Biotechnol Adv 17:143–182
Kumar A, Sharma N (2013) Protandrous protogynous dimorphism in indigenous selections from North Western India and some exotic cultivars of Persian walnut (Juglans regia L.). Adv Hortic Sci 61–66
Kuras A, Antonius K, Kalendar R, Kruczy´nska D, Korbin M (2013) Application of five DNA marker techniques to distinguish between five apple (Malus × domestica Borkh.) cultivars and their sports. J Hortic Sci Biotechnol 88(6):790–794
Li G, Quiros CF (2000) Use of amplified fragment length polymorphism markers for celery cultivar identification. HortScience 35(4):726–728
Mahmoodi R, Reza Dadpour M, Hassani D, Zeinalabedini M, Vendramin E, Micali S, Zaare NF (2019) Development of a core collection in Iranian walnut (Juglans regia L.) germplasm using the phenotypic diversity. Sci Hortic 249:439–448
Milovanov A, Zvyagin A, Daniyarov A, Kalendar R, Troshin L (2019) Genetic analysis of the grapevine genotypes of the Russian Vitis ampelographic collection using iPBS markers. Genetica 147(1):91–101
Muradoğlu F, Balta F, Battal P (2009) Endogenous hormone levels in bearing and non-bearing shoots of walnut (Juglans regia L.) and their mutual relationships. Acta Physiol Plant 32(1):53–57
Muradoğlu F, Oğuz Hİ, Yildiz K, Yilmaz H (2010) Some chemical composition of walnut (Juglans regia L.) selections from eastern Turkey. Afr J Agric Res 5(17):2379–2385
Naeem H, Awan FS, Dracatos PM, Sajid MW, Saleem S, Yousafi Q, Khan MS, Mehmood A, Zulfiqar B (2021) Population structure and phylogenetic relationship of Peach [Prunus persica (L.) Batsch] and Nectarine [Prunus persica var. nucipersica (L.) C.K. Schneid.] based on retrotransposon markers. Genet Resour Crop Evol 68:3011–3023
Nicese FP, Hormaza JI, McGranahan GH (1998) Molecular characterization and genetic relatedness among walnut (Juglans regia L.) genotypes based on RAPD markers. Euphytica 101:199–206
Orhan O, Eyduran SP, Poljuha D, Akin M, Weber T, Ercisli S (2020) Genetic diversity detection of seed-propagated walnut (Juglans regia L.) germplasm from Eastern Anatolia using SSR markers. Folia Hort 32(1):37–46
Ouyang Z, Wang Y, Ma T, Kanzana G, Wu F, Zhang J (2021) Genome-wide identification and development of LTR retrotransposon-based molecular markers for the Melilotus genus. Plants 10(5):890
Özer G, Bayraktar H, Baloch FS (2016) iPBS retrotransposons ‘A Universal Retrotransposons’ now in molecular phylogeny of fungal pathogens. Biochem Syst Ecol 68:142–147
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinform 28:2537–2539
Pérez-Vargas I, Portero Álvarez AM, Pérez de Paz PL, PÉrez JA (2020) Retrotransposon-based molecular markers as a tool in delimiting species in section Ryncholotus, a recent radiation group of Macaronesian Lotus. Syst Biodivers 19(2):110–120. https://doi.org/10.1080/14772000.2020.1827076
Poggetti L, Ermacora P, Cipriani G, Pavan F, Testolin R (2017) Morphological and carpological variability of walnut germplasm (Juglans regia L.) collected in North-Eastern Italy and selection of superior genotypes. Sci Hortic 225(18):615–619
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
R Core Team (2020) R: A language and environment for statistical computing. R found for stat comput, Vienna, Austria
Rahmani MS, Shabanian N, Khadivi-Khub A, Woeste KE, Badakhshan H, Alikhani L (2015) Population structure and genotypic variation of Crataegus pontica inferred by molecular markers. Gene 572(1):123–129. https://doi.org/10.1016/j.gene.2015.07.001
Rohlf FJ (2000) NTSYS-pc numerical taxonomy and multivariate analysis system. Version 2. 1. Exeter Software, Setauket.
Roldàn-Ruiz I, Dendauw J, Van Bockstaele E, Depicker A, De Loose MAFLP (2000) AFLP markers reveal high polymorphic rates in ryegrasses Lolium spp. Mol Breed 6:125–134
San Miguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Berhan-Melake A, Springer PS, Lee M, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768
Schulman AH, Flavell AJ, Ellis THN (2004) The application of LTR retrotransposons as molecular markers in plants. Methods Mol Biol 260:145–175
Sharma OC, Sharma SD (2001) Genetic divergence in seedling trees of Persian walnut (Juglans regia L.) for various metric nut and kernel characters in Himachal Pradesh. Sci Hortic 88:163–171
Skender A, Kurtovıc M, Drkenda P, Becırspahıc D, Ebrahımı A (2020) Phenotypic variability of autochthonous walnut (Juglans regia L.) genotypes in northwestern Bosnia and Herzegovina. Turk J Agric For 44:517–525
UPOV (2017) UPOV (International Union for the Protection of New Varieties of Plants), Draft guidelines for the conduct of tests for distinctness, uniformity and stability WALNUT).TG/125/7(proj.5).
Verma MK, Sharma VK, Lal S, Mır JI, Sofı AA, Sıngh DB, Pandıt AH, Mır MA, Mır A, Bhat HA (2020) Quality profiling of Indian walnut (Juglans regia) from Kashmir valley. Indian J Agric Sci 90(3):573–576
Wei T, Simko V (2017) R Package “corrplot” visualization of a correlation matrix (Version 0.84).
Wickham, H., 2016. ggplot2: Elegant graphics for data analysis. Springer, New York. ISBN 978-3-319 24277-4, https://ggplot2.tidyverse.org. https://doi.org/10.1007/978-3-319-24277-4
Yucel C, Baloch FS, Ozkan H (2009) Genetic analysis of some physical properties of bread wheat grain (Triticum aestivum L. em Thell). Turk J Agric For 33:525–535
Zhadan VM, Strukov MV (1977) Breeding walnut for fruit size. Plant Breed Abstract 47(11):918
Acknowledgements
Data of the manuscript were received from the Ph.D. thesis of İbrahim Başak
Author information
Authors and Affiliations
Contributions
İB performed field experiments. İB and FM carried out the laboratory studies. FM performed statistical analysis and data visualization. GÖ performed molecular studies. FM, GÖ, and İB wrote and reviewed the original draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare there is not any known financial or personal competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Başak, İ., Özer, G. & Muradoğlu, F. Morphometric traits and iPBS based molecular characterizations of walnut (Juglans regia L.) genotypes. Genet Resour Crop Evol 69, 2731–2743 (2022). https://doi.org/10.1007/s10722-022-01394-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01394-7