Abstract
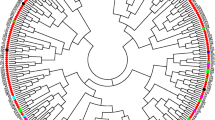
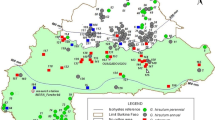
The remaining populations of perennial cotton (Gossypium hirsutum L. r. marie-galante Hutch.) that occur in northeastern Brazil are part of the primary gene pool for the creation of modern upland cotton cultivars. Therefore, we present fiber technological properties, genetic diversity and population structure analysis of four populations of G. hirsutum L. r. marie-galante from Paraíba state in Brazil, using the High-Volume Instrument (HVI) analytical system and Inter Simple Sequence Repeats (ISSRs) polymorphic loci, respectively. The fiber technological properties differed between populations. The average length, resistance, micronaire index, maturity index and count-strength-product of the populations are consistent with the fiber quality traits present in commercial upland cotton cultivars. The populations showed low levels of total (HT = 0.1305) and intrapopulation (HS = 0.0806) genetic diversity, besides high genetic differentiation (GST = 0.3824) and low indirect gene flow (Nm = 0.875) and are structured in two gene clusters by Bayesian inference. The positive relationship between genetic distance and geographical distance using the Mantel test (r = 0.8364; p value = 0.0038) suggests that genetic divergence among populations can be explained by spatial isolation. This information can support genetic conservation and the valuation of these populations for cotton breeding programs.


Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ayres M, Ayres Junior M, Ayres DL, Santos AS (2007) Bioestat 5.0 - Aplicações estatísticas nas áreas das ciências biomédicas. ONG Mamiraua, Belém
Bachelier B, Gourlot J (2018) A fibra do algodão. In: Belot J (ed). Manual da qualidade de fibra da AMPA. IMAmt, Cuiabá, pp. 28–57
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Brasil. Ministério da Agricultura, Pecuária e Abastecimento. Instrução Normativa nº 24, de 14 de julho de 2016. Regulamento Técnico do Algodão em Pluma. Diário Oficial da União, Brasília
Coêlho JD (2018) Produção de algodão Caderno setorial ETENE 2(3):1–7
Dongre AB, Bhandarkar M, Banerje S (2007) Genetic diversity in tetraploid and diploid cotton (Gossypium spp.) using ISSR and microsatellite DNA markers. Indian J Biotechnol 6:349–353
Doyle JJ, Doyle JL (1987) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, VonHoldt BM (2012) Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361. https://doi.org/10.1007/s12686-011-9548-7
Embrapa (2001) BRS 200 marron: cultivar de algodão de fibra colorida. Embrapa Algodão, Campina Grande
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14(8):2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Freire EC, Santos MSS, Medeiros LC, Andrade FP, Santos EO (1990) Avaliação preliminar da coleção de germoplasmas de algodoeiro arbóreo no nordeste do Brasil. Embrapa Algodão, Campina Grande
Freire EC, Costa JN, Andrade FP (1997) Recursos genéticos e melhoramento do algodão no Nordeste do Brasil. Simpósio sobre recursos genéticos no Nordeste, Petrolina. Accessed 23 May 2020
Giband M, Dessauw D, Barroso PAV (2010) Cotton: Taxonomy, origin and domestication. In: Wakelyn PJ, Chaudhry MR (eds) Cotton: technology for the 21st Century. International cotton advisory committee, Washington
Lewontin RC (1972) Testing the theory of natural selection. Nature 236:181–182
Mcdermott JM, Mcdonald BA (1993) Gene flow in plant pathosystem. Annu Rev Phytopathol 31:353–373
Medeiros AA, Canan B, Felipe MG, Bacurau MNV (2012) A cotonicultura no Estado do Rio Grande do Norte: aspectos históricos e perspectivas. Redige 3:1–17
Menezes IPP, Barroso PAV, Hoffmann LV, Lucena VS, Giband M (2010) Genetic diversity of mocó cotton (Gossypium hirsutum race marie galante) from the northeast of Brazil: implications for conservation. Botany 88(8):765–773. https://doi.org/10.1139/B10-045
Menezes IPP, Hoffmann LV, Barroso PAV (2015) Genetic characterization of cotton landraces found in the Paraíba and Rio Grande do Norte states. Crop Breed Appl Biotechnol 15(1):26–32. https://doi.org/10.1590/1984-70332015v15n1a4
Menezes IPP, Hoffmann LV, Lima TH, Silva AR, Lucena VS, Barroso PAV (2017) Genetic diversity of arboreal cotton populations of the Brazilian semiarid: a remnant primary gene pool for cotton cultivars. Genet Mol Res. https://doi.org/10.4238/gmr16039659
Moreira JAN, Beltrão NEM, Freire EC, Novaes Filho MB, Santos RF, Amorim Neto MS (1997) Decadência do algodoeiro Mocó e medidas para o seu soerguimento no Nordeste brasileiro. Embrapa Algodão, Campina Grande
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70(12):3321–3323. https://doi.org/10.1073/pnas.70.12.3321
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:90–583
Noormohammadi Y, Hasheminejad-Ahangarani F, Sheidai M, Ghasemzadeh-Baraki S (2013) Genetic diversity analysis in opal cotton hybrids based on SSR, ISSR, and RAPD markers. Genet Mol Res 12(1):256–269. https://doi.org/10.4238/2013.30.01.12
Paulino JS, Gomes RA (2015) Sementes da Paixão: agroecologia e resgate da tradição. Rev Econ Sociol Rural 53(3):517–528. https://doi.org/10.1590/1234-56781806-9479005303008
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Rocha GMG, Cavalcanti JJV, Carvalho LP, Santos RC (2016) Genetic divergence of colored cotton based on inter simple sequence repeat (ISSR) markers. Afr J Agric Res 11(29):2663–2668. https://doi.org/10.5897/AJAR2016.11074
Rodrigues F, Ribeiro MF (2014) Influence of experience on homing ability of foragers of Melipona mandacaia Smith (Hymenoptera: Apidae: Meliponini). Sociobiology 61(4): 523–528. https://doi.org/10.13102/sociobiology.v61i4.523-528
Rohlf FJ (2000) Numerical taxonomy and multivariate analysis system version 2.11. Applied Biostatistic, New York
Sheidai M, Noormohammadi Z, Shojaei-Jeshvaghani F, Ghsemzadeh-Baraki S (2012) Simple sequence repeat (SSR) and inter simple sequence repeat (ISSR) analyses of genetic diversity in tissue culture regenerated plants of cotton. Afr J Biotechnol 11(56):894–11900. https://doi.org/10.5897/AJB12.079
Silva AG, Pinto RS, Contrera FAL, Albuquerque PMC, Rêgo MMC (2014) Foraging distance of Melipona subnitida Ducke (Hymenoptera: Apidae). Sociobiology. 61 (4):494–501. doi: https://doi.org/10.13102/sociobiology.v61i4.494-501
Vidal Neto FC (2013) Freire EC (2013) Melhoramento genético do algodoeiro. In: Vidal Neto FC, Cavalcanti JJV (eds) Melhoramento genético de plantas no Nordeste. Embrapa, Brasília, DF, pp 49–83
Yeh FC, Boyle TYZ, Xiyan JM (1999) POPGENE version 131: Microsoft Window-based freeware for population genetic analysis. University of Alberta, Edmonton
Funding
Partial financial support has been received from Brazilian Agricultural Research Corporation and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001(PhD and postdoctoral scholarships).
Author information
Authors and Affiliations
Contributions
The authors Araújo, FS, Arriel, NH, Bruno, RLA and Medeiros, EP contributed to the study's conception and design. Material preparation, data collection and analysis were performed by Araújo, FS, Lima, LM and Souza, MA. The first draft of the manuscript was written by Araújo, FS and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose.
Consent for publication
All the authors approve the manuscript submission and agree to submit it to the Genetic Resources and Crop Evolution.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
dos Santos Araújo, F., Arriel, N.H.C., de Medeiros, E.P. et al. Multi-level characterization of perennial cotton (Gossypium hirsutum L. race marie-galante Hutch.) populations from the northeastern Brazil to the breeding and conservation of this germplasm. Genet Resour Crop Evol 69, 1219–1227 (2022). https://doi.org/10.1007/s10722-021-01299-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01299-x