Abstract
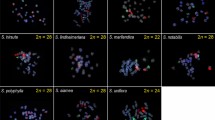
Most recent studies on Lilium species have been descriptive observations based on FISH (fluorescence in situ hybridization) analyses or phylogenetic studies focusing on only one section without taking the evolutionary framework of Lilium into account. To better understand the taxonomic status, evolution, and karyotype diversity of the genus Lilium, we conducted FISH mapping using 35S rDNA sequences in 29 important wild Lilium species covering four sections of the genus in China. Their molecular phylogenetic relationships were analyzed based on the internal transcribed spacer (ITS) region. For 35S rDNA, the hybridization loci always existed in pairs on homologous chromosomes. The number of hybridization signals varied from 4 to 14 in diploid species, and the signals were mainly located on the proximal region of the short arm of chromosomes. Among all the sections, sect. Sinomartagon had the most 35S rDNA signals. Genotype grouping based on 35S signal loci was largely consistent with groups in the phylogenetic tree constructed using ITS sequences, and different species in a sect./subsect. shared some common molecular and cytogenetic features. The results indicated that sect. Martagon is monophyletic in a single clade; Lophophorum should be treated as a different taxon from Sinomartagon; Lilium taliense and Lilium wardii Stapf ex Stern show close molecular and cytological phylogenetic relationships; Lilium duchartrei Franch. is more distantly related to other sect. Sinomartagon species; Lilium lophophorum (Bureau & Franch.) Franch. is distinct from other Lophophorum species; Leucolirion species form two relatively independent groups (6a and 6b, with Lilium henryi Baker and Lilium rosthornii Diels in 6a); and Nomocharis-like Lilium species should be treated as a different taxon, like sect. Lophophorum. Based on previous studies and our findings, we discuss the evolutionary trends of Lilium with references to its geographical distribution. We propose that sect. Lophophorum is a relatively primitive group, and species in subsect. Leucolirion 6b may be more evolved than those in 6a. These findings provide a new reference for the taxonomy, phylogeny, and breeding of Lilium.





Similar content being viewed by others
References
Ahn YJ, Hwang YJ, Younis A, Sung MS, Ramzan F, Kwon MJ et al (2017) Investigation of karyotypic composition and evolution in Lilium species belonging to the section Martagon. Plant Biotechnol Rep 11:407–416. https://doi.org/10.1007/s11816-017-0462-7
Arano H (1963) Analysis and phylogenetic considerations on Pertya and Ainsliaea. Bot Mag Tokyo 76:32–39
Cao QZ, Lian YQ, Wang LJ, Zhang Q et al (2019) Physical mapping of 45S rDNA loci in Lilium OT hybrids and interspecific hybrids with Lilium regale. Sci Hortic 252:48–54. https://doi.org/10.1016/j.scienta.2019.03.035
Comber HF (1949) A new classification of the genus Lilium. Lily Year Book RHS 13:86–105
De Jong PC (1974) Some notes on the evolution of lilies. N Am Lily Yearbook 27:23–28
Du YP, He HB, Wang ZX, Li S, Wei C, Yuan XN et al (2014a) Molecular phylogeny and genetic variation in the genus Lilium native to China based on the internal transcribed spacer sequences of nuclear ribosomal DNA. J Plant Res 127:249–263. https://doi.org/10.3389/Fpls.2017.01303
Du YP, Wei C, Wang ZX, Li S, He HB, Jia GX (2014b) Lilium spp. pollen in China (Liliaceae): taxonomic and phylogenetic implications and pollen evolution related to environmental conditions. PLoS ONE 9(1):e87841. https://doi.org/10.1371/journal.pone.0087841
Du YP, He HB, Wang ZX, Wei C, Li S, Jia GX (2014c) Investigation and evaluation of the genus Lilium resources native to China. Genet Resour Crop Evol 61(2):395–412. https://doi.org/10.1007/s10722-013-0045-6
Du F, Jiang J, Jia H, Zhao XY, Wang WH, Gao QK et al (2015) Selection of generally applicable SSR markers for evaluation of genetic diversity and identity in Lilium. Biochem Syst Ecol 61:278–285. https://doi.org/10.1016/j.bse.2015.05.002
Du YP, Bi Y, Zhang MF, Yang FP, Jia GX, Zhang XH (2017) Genome size diversity in Lilium (Liliaceae) is correlated with karyotype and environmental traits. Front Plant Sci 8:1303. https://doi.org/10.3389/Fpls.2017.01303
Dubouzet J, Shinoda K (1999a) ITS DNA sequence relationships between Lilium concolor Salisb., L. dauricum Ker-Gawl. and their putative hybrid, L. maculatum Thunb. Theor Appl Genet 98:213–218. https://doi.org/10.1007/s001220051060
Dubouzet J, Shinoda K (1999b) Phylogenetic analysis of the internal transcribed spacer region of Japanese Lilium species. Theor Appl Genet 98:954–960. https://doi.org/10.1007/s001220051155
El-Twab MHA, Abdel-Baset HD, Helmey RK, Atta AE-TM (2015) Characterization of chromosome complement in Achillea fragrantissima (Forsk) Schultz-Bip. analyzed by DAPI and FISH. Chromosome Bot 10:7–11. https://doi.org/10.3199/iscb.10.7
Gao YD, Gao XF (2016) Accommodating Nomocharis in Lilium (Liliaceae). Phytotaxa 277(2):205–210. https://doi.org/10.11646/phytotaxa.277.2.8
Gao YD, Zhou SD, He XJ (2011) Karyotype studies in thirty-two species of Lilium (Liliaceae) from China. Nord J Bot 29:746–761. https://doi.org/10.1111/j.1756-1051.2011.01069.x
Gao YD, Hohenegger M, Harris AJ, Zhou SD, He XJ, Wan J (2012) A new species in the genus Nomocharis Franchet (Liliaceae): evidence that brings the genus Nomocharis into Lilium. Plant Syst Evol 298:69–85. https://doi.org/10.1007/s00606-011-0524-1
Gao YD, Harris A, Zhou SD, He XJ (2013) Evolutionary events in Lilium (including Nomocharis, Liliaceae) are temporally correlated with orogenies of the Q–T plateau and the Hengduan Mountains. Mol Phylogenet Evol 68:443–460. https://doi.org/10.1016/j.ympev.2013.04.026
Gao T, Sun H, Fang L, Qian H, Xin H, Shi J, Wu Z, Xia M (2016) Cytogenetic analysis of Asiatic lily cultivars and their hybrids using fluorescence in situ hybridization. Acta Hortic 1027:177–184. https://doi.org/10.17660/ActaHortic.2014.1027.19
Garcia S, Kovařík A, Leitch AR, Garnatje T (2017) Cytogenetic features of rRNA genes across land plants: analysis of the Plant rDNA database. Plant J 89:1020–1030. https://doi.org/10.1111/tpj.13442
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp Ser 41:95–98
Harper ME, Saunders GF (1981) Localization of single copy DNA sequences on G-banded human chromosomes by in situ hybridization. Chromosoma 83:431–439. https://doi.org/10.1007/BF00327364
Haw SG (1986) The lilies of China. BT Batsford Ltd, London
Hayashi K, Kawano S (2000) Molecular systematics of Lilium and allied genera (Liliaceae): phylogenetic relationships among Lilium and related genera based on the rbcL and matK gene sequence data. Plant Spec Biol 15:73–93. https://doi.org/10.1046/j.1442-1984.2000.00025.x
Huang YF, Yang MX, Zhang H et al (2009) Genetic diversity and genetic structure analysis of the natural populations of Lilium brownii from Guangdong, China. Biochem Genet 47(7–8):503–510
Hwang YJ, Kim HH, Kim JB, Lim KB (2011) Karyotype analysis of Lilium tigrinum by FISH. Hortic Environ Biotechnol 52:292–297. https://doi.org/10.1007/s13580-011-0225-2
Hwang YJ, Song CM, Younis A, Kim CK, Kang YI, Lim KB (2015) Morphological characterization under different ecological habitats and physical mapping of 5S and 45S rDNA in Lilium distichum with fluorescence in situ hybridization. Rev Chilena de Historia Nat 88:8. https://doi.org/10.1186/s40693-015-0037-3
Jiang J, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49:1057–1068. https://doi.org/10.1139/g06-076
Lee JS, Lee PO, Lim YP, Shin EM, Park SY (1996) Classification of lilies using random amplified polymorphic DNA (RAPD) analysis. Acta Hort 414:137–144. https://doi.org/10.17660/ActaHortic.1996.414.15
Lee CS, Kim SC, Yeau SH, Lee NS (2011) Major lineages of the genus Lilium (Liliaceae) based on nrDNA ITS sequences, with special emphasis on the Korean species. J Plant Biol 54:159–171. https://doi.org/10.1007/s12374-011-9152-0
Lee H, Younis A, Hwang YJ, Kang YI, Lim KB (2014) Molecular cytogenetic analysis and phylogenetic relationship of 5S and 45S ribosomal DNA in sinomartagon Lilium species by fluorescence in situ hybridization (FISH). Hortic Environ Biotechnol 55:514–523. https://doi.org/10.1007/s13580-014-0066-x
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
Liang SY, Zhang WX (1985) Pollen morphology of the genus Nomocharis and its delimitation with Lilium. Acta Phytotax Sin 23:405–417 (In Chinese)
Lim KB (2014) Ecological morphological and cytogenetic analysis of Korean Martagon Lilium species. Acta Hort 1027:41–46
Lim KB, Chung JD, Van Kronenburg BC, Ramanna MS, De Jong JH, Van Tuyl JM (2000) Introgression of Lilium rubellum Baker chromosomes into L. longiflorum Thunb.: a genome painting study of the F1 hybrid, BC1 and BC2 progenies. Chromosome Res 8:119–125. https://doi.org/10.1023/A:1009290418889
Lim KB, Wennekes J, De Jong JH, Jacobsen E, Van Tuyl JM (2001) Karyotype analysis of Lilium longiflorum and Lilium rubellum by chromosome banding and fluorescence in situ hybridisation. Genome 44(5):911–918. https://doi.org/10.1139/g01-066
Liu GX, Zhang XL, Lan Y, Xin H, Hu F, Wu Z, Shi J, Xi ML (2017) Karyotype and fluorescence in situ hybridization analysis of 15 Lilium species from China. J Am Soc Horticult Sci 142:298–305. https://doi.org/10.21273/Jashs04094-17
Marasek A, Hasterok R, Wiejacha K, Orlikowska T (2004) Determination by GISH and FISH of hybrid status in Lilium. Hereditas 140(1):1–7. https://doi.org/10.1111/j.1601-5223.2004.01721.x
Muratović E (2010) Karyotype evolution and speciation of European lilies from Lilium sect. Liriotypus. Taxon 59:165–175. https://doi.org/10.2307/27757060
Nishikawa T, Okazaki K, Uchino T, Arakawa K, Nagamine T (1999) A molecular phylogeny of Lilium in the internal transcribed spacer region of nuclear ribosomal DNA. J Mol Evol 49:238–249. https://doi.org/10.1007/PL00006546
Nishikawa T, Okazaki K, Arakawa K, Nagamine S (2001) Phylogenetic analysis of section Sinomartagon in genus Lilium using sequences of the internal transcribed spacer region in nuclear ribosomal DNA. Breed Sci 51:39–46
Page RMD (1996) Treeview: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Paszko B (2006) A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol 258:39–48. https://doi.org/10.1007/s00606-005-0389-2
Peruzzi L, Leitch IJ, Caparelli KF (2009) Chromosome diversity and evolution in Liliaceae. Ann Bot 103:459–475. https://doi.org/10.1093/aob/mcn230
Ren GL, Zhang XX, Zhou SJ (2012) Hybridization and identification between Asiatic lily and Lilium tsingtauense using FISH. Acta Horticult Sin 39:588–592 (In Chinese)
Roa F, Guerra M (2012) Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evol Biol 12:225. https://doi.org/10.1186/1471-2148-12-225
Siljak-Yakovlev S, Peccenini S, Muratovic E, Zoldos V, Robin O, Vallès SJ (2003) Chromosomal differentiation and genome size in three European mountain Lilium species. Plant Syst Evol 236:165. https://doi.org/10.1007/s00606-002-0240-y
Smyth DR, Kongsuwan K, Wisudharomn S (1989) A Survey of C-Band Patterns in Chromosomes of Lilium (Liliaceae). Plant Syst Evol 163:53–69. https://doi.org/10.1007/Bf00936153
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stebbins GL (1971) Chromosomal evolution in higher plants. Edward Arnold, pp 71–100
Stewart RN (1947) The morphology of somatic chromosomes in Lilium. Am J Bot 34:9–26. https://doi.org/10.2307/2437190
Sultana S, Lee SH, Bang JW, Choi HW (2010) Physical mapping of rRNA gene loci and inter-specific relationships in wild Lilium distributed in Korea. J Plant Biol 53:433–443. https://doi.org/10.1007/s12374-010-9133-8
Swofford DL (2000) PAUP* 4.0: phylogenetic analysis using parsimony (* and other methods). Version 4.0 b10. Sinauer Associates, Sunderland
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. https://doi.org/10.1093/nar/25.24.4876
Wang FZ, Tang J (1980) Lilium L. Flora Reipublicae Popularis Sinicae, vol 14. Science Press, Beijing, pp 116–157
Younis A, Ramzan F, Hwang YJ, Lim KB (2015) FISH and GISH: molecular cytogenetic tools and their applications in ornamental plants. Plant Cell Rep 34:1477–1488. https://doi.org/10.1007/s00299-015-1828-3
Zhou SJ, Van Tuyl J, Zhang DK, Xia YP, Li F (2008) Physical localization of 45S rDNA on the chromosomes of 4 species of the genus Lilium. Acta Horticult Sin 35:859–862 (In Chinese)
Zhou SJ, Zhong L, Zhang L, Xu ZH, Liu XX, Li KH, Zhou GX (2015) Study on the homology of the genomes of tetraploid Asiatic lilies (Lilium) using FISH. Genome 58(11):453–461. https://doi.org/10.1139/gen-2015-0057
Acknowledgements
This work was supported by National Key R&D Program of China (Grant No.2019YFD1000403) and the National Natural Science Foundation of China (31772348).
Author information
Authors and Affiliations
Contributions
Y.-P. Zhou, Zh.-X. Wang, and G.-X. Jia designed experiments; Y.-P. Zhou, Zh.-X. Wang, and J.-W. Li carried out experiments and analyzed experimental results. The manuscript was written by Y.-P. Zhou and Zh.-X. Wang and modified by Y.-P. Zhou, Y.-P. Du, and H.-B. He.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhou, YP., Wang, ZX., Du, YP. et al. Fluorescence in situ hybridization of 35S rDNA sites and karyotype of wild Lilium (Liliaceae) species from China: taxonomic and phylogenetic implications. Genet Resour Crop Evol 67, 1601–1617 (2020). https://doi.org/10.1007/s10722-020-00936-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-020-00936-1