Abstract
The paper reports on the study of DNA methylation patterns in chromosomes of an octoploid selection obtained from an amphiploid Avena barbata × Avena sativa ssp. nuda, their alteration and plant structural changes induced by 5-azacytidine (5-azaC) treatment. Subtelomere/telomere regions and some chromosomes remained heavily methylated after this practise. The increased concentration of 5-azaC strengthened the demethylation process. NOR rDNA loci on five and 5S rDNA doubled loci on three pairs of chromosomes were detected. Some 5S loci were colocalised with NORs. The number of active NORs was increased by one homologous pair after demethylation and this was related to minor loci. The NORs were mainly located on A/D genomes. Patterns of nucleoli fusion varied between main and lateral roots and demethylation increased the number of uninucleolar nuclei. Seed germination was inhibited and plant growth decreased under demethylation. 5-azaC also induced distinct anomalies in endosperm development. Extensive parts of the tissue were composed of defected cells or remained empty. Polyploidised clones of starchy or aleurone cells were observed, as were aleurone cells exhibiting new rare phenotypes.
Similar content being viewed by others
Introduction
The number of heterozygous chromosomal rearrangements is an important factor causing hybrid pollen sterility. This defect in the biology of a species disappears after natural or artificial chromosome doubling, when amphiploids are created. In nature such polyploids are more common than autopolyploids (Grant 1981). In the genus Avena L., frequent translocations occurred during its natural polyploidisation process (Holden 1966). Nishiyama (1962) added an artificial pentaploid hybrid Avena barbata Pott ex Link × A. sativa subsp. nuda (L.) Gillet et Magne to variation existing in oat. This hybrid was highly sterile. Decaploid amphiploids were obtained after application of colchicine. These decaploids were variable in breeding behaviour; however, selection in successive generations provided types expressing higher fertility and stability of characters. Kushwaha et al. (2004) observed cytogenetic instability and loss of chromosomes in A14–16 oat amphiploids derived from pentaploid hybrids (6x/4x). Some stable polysomics were selected, always with the chromosome number of the maternal parent or higher. Also, a decaploid (2n = 70) was obtained with 88.1–94.2% of bivalents. Further experiments conducted by Nishiyama et al. (1963) proved that oat decaploids evolved to lower levels of polyploidy, 8x and 6x, which were more stable.
Parental genomes of alloploids are also variable in the molecular sense. This dynamic appeared in grass (cereals) allopolyploids in the form of changes of genome methylation patterns, rapid epigenetic gene silencing and activation of retrotransposons or nucleolar dominance (Liu and Wendel 2003).
In eukaryots, methylation sites are not randomly scattered among and along chromosomes (Antequera and Bird 1993; Siroky et al. 1998). Dotted methylation signals have been observed along all Triticale chromosomes (Castilho et al. 1999). Methylation of heterochromatic regions is common (Ruffini Castiglione et al. 2002); therefore, one can expect that genome C in Avena sativa, which is highly heterochromatic (Jellen et al. 1993), will be methylated. Also, subtelomeric and telomeric parts of chromosomes are methylated and react to demethylating drugs (Majerová et al. 2011).
Loci of rDNA are often non-randomly methylated and in plant hybrids this phenomenon is documented as nucleolar dominance of one parent (Pikaard 2000). In young wheat-rye amphiploids, the alterations in cytosine methylation occurred in 13% of the genomic loci. Such methylation suppressed the rye rDNA loci. These loci were activated after application of 5-aza-cytidine (Castilho et al. 1999; Levy and Feldman 2002).
NOR-rDNA loci are mostly classified as major and minor. In common wheat, the smallest locus is located on chromosome 1A and the major one on chromosome 6B (Lacadena et al. 1988). In hybrid progeny of wheat × Thinopyrum distichum (Thunb.) Á. Löve, NORs of Thinopyrum Á. Löve origin are usually not expressed, especially a tiny pair located terminally (Armstrong et al. 1991; Kosina and Heslop-Harrison 1996). In tetraploid oat Avena barbata four pairs of NORs have been determined, but only two major NORs are expressed as Ag-NOR. It is probable that minor loci determined on chromosomes from the B genome have been suppressed (Irigoyen et al. 2001; Florek 2013). Badaeva et al. (2011) recognised as many as eight minor NORs in oat hexaploids. Variation between the expression of major and minor loci in various organs or tissues with native or demethylated genomes seems to be important during the development of the plant.
The level of methylation varies, for instance, from ca 5% of cytosines in Arabidopsis to 20% in wheat, and this depends on the amount of repetitive DNA. DNA methylation is required for normal plant development and chromosome stability (Gehring and Henikoff 2007). In addition, the level of methylation is tissue-specific and depends on the degree of tissue differentiation (Messeguer et al. 1991). The tissue in which methylation-demethylation plays an important role is endosperm. The Arabidopsis endosperm with its many silenced genes is an example of this (Hsieh et al. 2009). In rice, the expression of genes responsible for the synthesis of storage proteins and starch in endosperm is activated by hypomethylation (Zemach et al. 2010). The endosperm is a tissue, where maternal and paternal genomes interact in different doses. A hybrid alloploid endosperm seems to be a more profitable subject for the study of methylation/demethylation effects due to the suppression of one of the parental genomes as a whole. This phenomenon adds a new variation of the tissue when compared to that in a pure species. This has been proved in the endosperm of various oat amphiploids in which the A genome dominates over the B genome (Tomaszewska and Kosina 2013). Analysis of endosperm mutants in Arabidopsis showed that change of methylation patterns can relate to hundreds of genes (Grossniklaus et al. 2001). Such a quantitative Arabidopsis endosperm epigenetics could be further complicated by the loss of chromosomes of different genomes in the complex Avena amphiploids (Tomaszewska and Kosina 2015).
Materials and methods
Plant material
Two parental species, a maternal wild tetraploid species Avena barbata Pott ex Link (2n = 4x = 28, AABB genomes) and a paternal cultivated hexaploid species A. sativa subsp. nuda (L.) Gillet et Magne (2n = 6x = 42, AACCDD genomes), were crossed and a pentaploid F1 hybrid (2n = 5x = 35, AABCD genomes) was obtained (Nishiyama 1962). A decaploid synthetic oat (2n = 10x = 70, AAAABBCCDD genomes) has been obtained by doubling chromosome numbers of a pentaploid hybrid. Then, an octoploid oat (2n = 8x = 56), with an unknown set of genomes, was selected from the artificial decaploid and this octoploid as the accession Clav 7903 has been received from the National Small Grains Collection in USA, and was used in this study. Ripe caryopses for cytogenetic and structural studies were taken from plants cultivated on small plots or in pots, in the grass collection maintained by R. Kosina. During the plot or pot experiments plant were grown under the same soil-climatic conditions. So, the study material was treated as originating from a completely randomised one-way classification design.
Chromosome preparation
For cytogenetic and caryopsis anatomy analyses, seeds were germinated at 24 °C for 2–4 days in Petri dishes containing either tap water or 0.5 mM solution of 5-azacytidine (5-azaC solution was freshly prepared every day). Roots 1–2 cm in length were immediately immersed in ice-cold water and incubated for 24 h in small glass tubes. Then, the roots were immediately fixed in a fresh ethanol: glacial acetic acid (3:1) mixture for 2–10 h at room temperature and the material was stored at −20 °C until required. Chromosome preparations were made according to Schwarzacher and Heslop-Harisson (2000).
Silver staining
Silver staining was followed a technique described by Friebe et al. (1996). Slides were treated with a borate buffer (pH 9.2) for 15–30 min and a few drops of freshly prepared 50% silver nitrate in distilled water was applied to each preparation. Slides were covered with a nylon mesh and incubated in a humid chamber at 45 °C for 1–2 h, washed in distilled water and air-dried. The number of active rDNA loci was determined and chromosome pictures were taken. Then, the silver staining was removed and slides were again used for fluorescence in situ hybridisation (FISH).
DNA probes
GISH and FISH signals were obtained with the use of the following probes, according to Schwarzacher and Heslop-Harisson (2000):
-
1.
Total genomic DNA was isolated in 2% CTAB (hexadecyltrimethylammonium bromide) isolation buffer from leaves of Avena nuda L. (AA genomes) according to the protocol of Doyle and Doyle (1987) and then used as a probe in GISH analyses. This probe was labelled with DIG (digoxigenin-11-dUTP, Roche, 11093088910) using a nick translation kit according to the manufacturer’s protocol (Roche, 10976776001).
-
2.
25S rDNA 2.3 kb ClaI fragment of the 25S rDNA ribosomal gene from Arabidopsis thaliana (L.) Heynh. (Unfried and Gruendler 1990) was labelled with DIG (digoxigenin-11-dUTP, Roche, 11093088910) using a nick translation kit according to the manufacturer’s protocol (Roche, 10976776001).
-
3.
pTa794 410 bp fragment of 5S rDNA unit isolated from Triticum aestivum L. (Gerlach and Dyer 1980) was labelled with TRITC (tetramethyl-rhodamine-5-dUTP, Roche 11534378910). The oligonucleotides and conditions used for PCR labelling were as follows: universal M13 forward (5′-CAG GGT TTT CCC AGT CAC GA-3′) and reverse (5′-CGG ATA ACA ATT TCA CAC AGG A-3′) sequencing primers.
In situ hybridisation (GISH, FISH)
After screening under a phase-contrast microscope, only slides with well-spread chromosomes were pretreated with DNase-free RNase A for 1 h at 37 °C, then washed three times in a 2× SSC buffer for 5 min per slide and post-fixed in 4% paraformaldehyde in a 1× PBS buffer (0.01 M sodium phosphate and 0.14 M NaCl, pH 7.0) for 10 min, washed in a 2× SSC buffer for 15 min, dehydrated in an ethanol series (70, 90 and 100%) and air-dried. All DNA probes were mixed to a concentration of about 100 ng per slide along with 50% deionised formamide, 10% dextran sulphate, 2× SSC, 0.5% SDS and sonicated salmon sperm blocking DNA, 50× probe (Sigma, cat. no. D-9156). The hybridisation mix was denaturated at 70 °C for 5 min and placed on ice for 10 min. Chromosome preparations and predenaturated probes were denaturated together on a hot plate (Hybaid Thermal Cycler PCR-in situ) at 75 °C for 5–10 min and then incubated for 12–16 h at 37 °C in a humid chamber. After hybridisation, slides were washed for 2 × 5 min in 15% formamide in 0.1× SSC at 42 °C followed by several washes in a 2× SSC buffer at 42 °C for 3 min and 2 × 3 min in 2× SSC at room temperature. After blocking in a detection buffer (see the protocol of Jenkins and Hasterok 2007) digoxigenated probes were immunodetected by antidigoxigenin antibodies conjugated with fluorescein isothiocyanate (FITC, Roche, 11207741910) or antidigoxigenin antibodies conjugated with rhodamine (Roche, 11207750910) in the same buffer (antibody dilution 1:12). Preparations were counterstained with DAPI (4′,6-diamino-2-phenylindole) and mounted in the antifade buffer (Citifluor). Cytogenetic slides were documented by using different epifluorescence microscopes: an Olympus BX50 with a DP71 digital camera and CellB software; an Olympus Provis AXE with a CCD C5810 Hamamatsu Inc. digital camera; an Olympus BX60 with an Olympus E-520 camera. The microscopy data were processed using ImageJ software.
Immunodetection of 5-methylocytosine (5-meC)
Slides were denaturated in 70% formamide for 2 min at 70 °C and blocked with 2% BSA for 10 min at 25 °C. Methylated cytosines were immunodetected with mouse monoclonal antibodies [33D3] raised against 5-mC [Abcam, ab 10805; 1:100 in 2% bovine serum albumin (BSA) in 1× phosphate-buffered saline (PBS)] and goat anti-mouse secondary antibody conjugated with Alexa 488 (Invitrogen, 1:250 in 2% BSA in 1 × PBS). Incubation with primary antibody was performed at 25 °C for 1 h and after washing in 1× PBS, the secondary antibody was applied under the same conditions. Chromosomes were counterstained with DAPI in Vectashield (Vector Laboratories). Slides were observed in an Axioskop 20 epifluorescence microscope (Zeiss) and pictures were taken with an AxioCam HRC camera. Pictures were processed with ImageJ 1.46 software (Wayne Resband National Institutes of Health, USA) and presented in Excel in the form of scanning diagrams. The whole cytogenetic study was made for random samples n ≥ 100.
Multivariate nucleolar behaviour data were elaborated with numerical taxonomy software and presented in the form of a minimum scanning tree in an ordination space obtained using non-metric multidimensional scaling (Rohlf 1994).
Morphology of caryopsis
For caryopsis anatomy analyses, seeds were germinated at 24 °C for 2–4 days in Petri dishes containing either tap water for control or 0.5 mM solution of 5-azacytidine for demethylation (5-azaC solution was freshly prepared every day). Ripe caryopses were sampled from the mature plants. Caryopses were fixed in FAA (40% formaldehyde—50% ethanol—10% glacial acetic acid). Then, they were rinsed several times in distilled water and cut with a freezing microtome K TS-II (an USSR equipment). Cross-sections of caryopses about 40 µm in thickness were mounted in glycerine as semi-permanent slides. From fixed caryopses the aleurone layer was also isolated and its surface was studied. The slides were documented in a polarising Amplival microscope (Carl Zeiss, Jena, Germany). Images were taken with an Olympus E-520 camera (Olympus Imaging Europa GMBH, Hamburg, Germany).
All the structural observations (germination, plant growth, caryopsis sections) were conducted on random samples n ≥ 60
Results and discussion
Demethylation by 5-azacytidine
Detection of methylated cytosines in root metaphase chromosomes and interphase nuclei showed that all genomes of the amphiploid were evenly and highly methylated (Fig. 1A-C). A high correlation exists between DAPI (blue) and Alexa 488 (green) fluorescence (Fig. 1C). After application of 5-azaC (0.5 mM), the level of methylated cytosines versus DAPI fluorescence of the total DNA was decreased (Fig. 2A-C). Only some scattered points in a nuclear space were still highly methylated (Fig. 2C). The arithmetic averages of pixel fluorescence, calculated for two hundred interphase nuclei, for native methylated genomes and demethylated ones are 44.5 and 8.2, respectively. The Student’s t test for unequal variances showed that the difference between both averages is statistically significant for P = 0.01. Application of a higher concentration of 5-azaC (1.0 mM) resulted in a higher level of demethylation and fewer points of green fluorescence were visible in interphase nuclei (Fig. 2D, E). These points relate, among others, to telomeric/subtelomeric methylated regions. Patterns of demethylation of metaphase chromosomes (Fig. 3) show that subtelomeric/telomeric parts were heavily methylated and remained undemethylated after 5-azaC treatment. Some chromosomes displayed intercalary sites resistant to 5-azaC, others remained methylated as a whole, in some sites very heavily. Demethylation removed 5-methylcytosine from large parts of the genomes (compare Figs. 1, 2, 3).
In situ immunodetection of 5-methylocytosine (5-meC) residues in a root interphase nucleus in water germinated seeds of the Avena amphiploid with methylated native genomes. A DAPI fluorescence, B Alexa 488 fluorescence, C diagram of the intensity (y axis) of DAPI (black line) and Alexa 488 (dotted line) fluorescence signals across the same nucleus in A and B (x axis, µm), respectively
In situ immunodetection of 5-methylocytosine (5-meC) residues in a root interphase nucleus of germinated seeds of the Avena amphiploid after 0.5 mM 5-azaC treatment. A DAPI fluorescence, B Alexa 488 fluorescence, C diagram of the intensity (y axis) of DAPI (black line) and Alexa 488 (dotted line) fluorescence signals across the same nucleus in A and B (x axis, µm), respectively. D, E for another nucleus—the same as for A and B, respectively, for 1.0 mM 5-azaC treatment
In situ immunodetection of 5-methylocytosine (5-meC) residues in a root metaphase in germinated seeds of the Avena amphiploid after 0.5 mM 5-azaC treatment (A DAPI fluorescence; B Alexa 488 fluorescence, bold arrows—5-meC residues in subtelomeres/telomeres, arrowheads for intercalar positions of residues, white arrows for chromosome arms (heterochromatic) heavily metylated and thin arrows for hypomethylated arms
Antequera and Bird (1993) discussed the distribution of heavily methylated CG domains in eukaryotic genomes. Numerous methylated domains are situated in an inert DNA and regions containing transposons, others unmethylated CpGs are clustered into islands in transcripted active genes. This means that methylation is non-randomly scattered along chromosomes. Sites with high and low levels of methylation along chromosomes have been detected in a hexaploid Triticale. Demethylation with 5-azacytidine induced chromosome breakage and translocations and formation of micronuclei of rye origin (Castilho et al. 1999). On the other hand, the level of translocations does not differ in native and demethylated by 5-azaC genomes of the Avena amphiploid (Florek 2013). Demethylation appeared to be 5-azaC concentration dependent in Brachypodium distachyon (L.) P. Beauv. Chromosomes have been demethylated at higher concentrations (0.1 mmol/L), but not so clearly at lower concentrations. Differences were noted in the hypomethylation level between homologous chromosomes, along chromosomes and between non-homologous ones (Borowska et al. 2011). Similarly, distinct differences in hypomethylation pattern are visible between chromosomes in the Avena amphiploid (see various arrows in Fig. 3B). Lewis and Bird (1991) related the methylation level to heterochromatinisation of chromosomes. Highly heterochromatic chromosomes from the C genome in oat hexaploids (AACCDD) (Phillips et al. 1995) are probably those heavily methylated. The short arms of these chromosomes are more heterochromatinised (methylated) than long ones and this difference is marked in Fig. 3B (normal and thin arrows). In the Avena amphiploid, higher concentrations of 5-azaC (0.5 vs. 1.0 mM) demethylated oat chromosomes more.
Methylation patterns observed in different plant organs are evidently correlated with normal development (Heslop-Harrison 2000; Gehring and Henikoff 2007). In endosperm tissue demethylation must be done to enable the expression of many previously imprinted genes (Hsieh et al. 2009). Plant telomeres are also evidently methylated, but heterochromatin patterns and its subtelomeric and telomeric methylation are inter-specifically variable (Ruffini Castiglione et al. 2002). For instance, telomeres are not hypermethylated in Melandrium album (Siroky et al. 1998), while they are heavily methylated in a grass Zingeria biebersteiniana (Claus) P.A. Smirn. (Cremonini et al. 2003). A careful interpretation of Fig. 3E, G presented in Castilho et al.’s (1999) paper shows that telomeric/subtelomeric regions are distinctly methylated in Triticale, but after 5-azaC treatment methylation of telomeres persisted only in a few cases. In the Avena amphiploid, terminal regions of some chromosomes maintained strong signals of methylation, while others were significantly demethylated by 5-azaC. Heavy methylation, despite demethylation by 5-azaC, has been visible along some chromosomes of the amphiploid and these chromosomes belong probably to the C genome, which have proved to be highly heterochromatic (Phillips et al. 1995; Jellen et al. 1993).
rDNA cytogenetics
rDNA signals and their activity
Metaphase FISH detection for 25S (green, and marked by solid lines) and 5S (red, and marked by dotted lines) rDNA revealed 5 and 6 loci per haploid set of chromosomes, respectively (Fig. 4A). The 5S loci are localised as subterminal-terminal plus interstitial on three chromosomes of the haploid set (Fig. 4B, C). The 25S loci are always subterminally or terminally localised and on three chromosomes per haploid set of genomes they are colocalized with 5S signals. On two chromosomes of the haploid set, the rDNA signals relate only to 25S genes and one is minor (Fig. 4B, C). The 25S genes are detected on five pairs of chromosomes, while 5S genes are doubled on three pairs.
Fluorescence of 5S (red and shown by dotted lines) and 25S (green and shown by solid lines) rDNA in root metaphase chromosomes stained with DAPI in the Avena amphiploid with methylated, native genomes. A Metaphase, white arrows for a pair of minor 25S loci; B ordered chromosomes with detection of rDNA loci; C a chromosome rDNA loci ideogram. (Color figure online)
On metaphase chromosomes, eight SAT-chromosomes were detected in A/D genomes (green or lighter) and two pairs of SATs, including one translocated pair, were noted for genomes of B/C origin (red or darker, Fig. 5). This proves that the main activity of NOR-rDNA is expressed on A and/or D genomes (major loci), and C and/or B genomes seem to be less active in this respect. In Fig. 5 several green–red (lighter–darker) translocations between A/D and B/C genomes origin can be identified (see white arrows and dotted lines).
GISH of SAT-chromosomes of the Avena amphiploid with methylated, native genomes (the A genome probe highlighted in yellow, lighter). Eight NORs on yellow (lighter) A/D genomes (dotted circles), one pair on red (darker) C/B genomes (thick circles) and two translocated NORs of C/B origin (thin circles). Several red-yellow (darker-lighter) tanslocations, probably of A/C genomes origin, are marked (white arrows and dotted lines). (Color figure online)
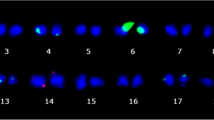
5-azaC treatment activated one additional pair of 25S rDNA versus the rDNA loci in native methylated metaphases (Fig. 6). Six active and four non-active nucleolar loci (Ag-NORs) were observed in native metaphases (Fig. 6A, B; see white and black arrows), while in demethylated metaphases eight active and two non-active loci were detected (Fig. 6C, D). Non-active loci expressed weaker signals of fluorescence.
FISH for rDNA loci (red for 5S and green for 25S) on metaphase chromosomes (A, C counterstained by DAPI) and the same metaphases (B, D respectively) with Ag-NOR loci in the Avena amphiploid. White arrows in A, B show 25S rDNA loci which are not active in the form of NORs; black arrows in C, D show 25S rDNA loci active as Ag-NORs. (Color figure online)
Florek (2013) detected nucleolar rDNAs on four pairs of chromosomes in Avena barbata (AABB genomes). The 5S loci were doubled and colocalised with the nucleolar rDNA on two of these four pairs. The same picture was described by Katsiotis et al. (1997) for A. vaviloviana (AABB). In A. barbata, all loci of both kinds of rDNA were equally expressed on A and B genomes, but one pair of NOR-rDNA loci on the genome B not colocalized with 5S was weaker (Irigoyen et al. 2001).
In other oat tetraploids (A. maroccana Gand., A. murphyi Ladiz.), four NORs have also been identified and localised on chromosomes from the A genome. Six 5S rDNA loci have been detected on the same genome, four of them are colocalised with the nucleolar ones. Oat hexaploids exhibit six nucleolar loci (four on A and two on D genome) plus eight (!) very weak ones, and there are also twelve 5S loci on A and D genomes, and some of these are colocalised (Linares et al. 1996).
Nucleolar rDNA sequences were detected by Jellen et al. (1994b) on six SAT-chromosomes in Avena sativa (AACCDD genomes). Slightly different data were presented by Badaeva et al. (2011) for various hexaploid oats with AACCDD genomes. Three major and four minor NORs and six signals for 5S rRNA were detected per haploid genome. Two 5S rDNA loci were colocalised with NORs. All rDNA loci were mapped on chromosomes belonging to A and D genomes, on which the positions of 5S and 45S rDNA loci were almost identical. Yang et al. (1999) detected all NOR-rDNA loci on A/D genomes in Avena fatua L. (AACCDD) and assumed that some minor ones were on the C genome. In addition, Nikoloudakis and Katsiotis (2008) found six major and six minor sites of nucleolar rDNA on chromosomes of A/D genomes in Avena sativa (AACCDD) and two minor signals of rDNA were observed on the C genome in A. fatua. Minor rDNA loci were also characteristic for the C genome in other oat species. Thus, detection of minor loci of rDNA may depend on the sensitivity of the protocol used for in situ hybridisation. In the Avena amphiploid, one pair of NOR-rDNA was minor (see Fig. 4b). In total, eight SAT-chromosomes in the amphiploid belong to A/D genomes. AAAA genomes originate from both parents. Two pairs of NOR probably belong to B or C genomes, including one translocated pair. It is also highly probable that translocations documented in the amphiploid occurred between A/D and C genomes (see Fig. 5; Florek 2013). The observed instability of the number of amphiploid chromosomes (Florek 2013) is related to the loss of some chromosomes from the B genome for hyperoctoploids, and from the C genome for hypooctoploids. Thus, the genomic set of the octoploid amphiploid could be AAAADDCC.
The 5S rDNA loci versus NORs can be colocalised, adjacent, neighbouring or located on different chromosomes. Spatial relationships on chromosomes of both kinds of rDNA are the same in two tetraploid oats, Avena vaviloviana (Malzev) Mordv. and A. barbata with AABB genomes. Eight 5S rDNA loci are colocalised either with NORs or are on other arms of the same chromosomes (Katsiotis et al. 1997; Florek 2013).
The colocalised positions of 5S and NORs have also been documented in A. murphyi (AACC) and A. sativa (AACCDD). A chromosome from the C genome (A. clauda Dur.) presents 5S and 26S rDNA at sites distant from each other on the same arm. Such a chromosome has not been noted in the Avena amphiploid. Some 5S loci are very weak and in A. sativa terminal translocations change their positions (Linares et al. 1996). Similar terminal translocations on chromosomes belonging to the A genome have been documented in the Avena amphiploid (Florek 2013) and they are of the A/C type characteristic for A. sativa, because no translocations between A and B genomes have been detected in A. barbata (Irigoyen et al. 2001; Florek 2013). It has been proved that all oat hexaploids are cytogenetically close to each other and differences between them are caused by translocations. In Avena sativa 5S and 26S rDNA loci are colocalised on chromosomes from A and D genomes, some weaker ones have been detected on the C genome (Badaeva et al. 2011). Two chromosomes, detected by Badaeva et al.’s as IA (A genome) and ID (D genome), show the same location of rDNAs sites as in the chromosome diagram for the Avena amphiploid (Fig. 4) and at least one pair of chromosomes from the B genome in A. barbata is similar (Irigoyen et al. 2001). This means that the diagram (Fig. 4) can present rDNA chromosomes from A and D, and some from B genome. A set of chromosomes which remained heavily methylated after 5-azaC application are related to heterochromatic chromosomes (Florek 2013), which can belong to the heterochromatic C genome. The level of reciprocal translocations between A and C genomes (Chen and Armstrong 1994; Jellen et al. 1994a), which can also be interpreted from the data presented in Fig. 5 and by reinterpretation of Florek’s data (Florek 2013), and the presence of C genome in the Avena amphiploid, allow it to be stated that genome B appeared more unstable and only some its chromosomes can be present in the hyperoctoploid lines.
Nucleoli
The number of nucleoli (Ag-NORs) per nucleus was counted for main and lateral roots with methylated and demethylated genomes (Fig. 7A, B). Nucleoli are commonly fused during the cell cycle. In the main roots, merging of a maximum of eight nucleoli, most often into one or two per nucleus was observed after the 5-azaC treatment, and this process is more intensive than that seen in the untreated main roots (Fig. 7A). The modal value of the number of nucleoli in demethylated nuclei is 2, while for native, methylated ones it is 3. In the lateral roots, merging of nucleoli into one or two bodies is more frequent (Fig. 7B). In both root types, the modal number of nucleoli per nucleus after demethylation is 2. The distribution of the nucleoli number is skewed and more leptocurtic for the lateral roots. Both diagrams (Fig. 7) show that the 5-azaC treatment differentiated a nucleolar fusion pattern between two root types.
Cytogenetic OTUs such as lrd, mrd, lrc and mrc (mr—main roots, lr—lateral roots, d—demethylation, c—control), characterised by patterns of variation of the nucleoli numbers, are, after the application of non-metric multidimensional scaling, positioned in an ordination space between two arbitrary accepted nucleolar states: “uninucleolar”, that is, fusion of all nucleoli into one nucleolus; and “anucleolar”, that is, fragmentation of all nucleoli up to their disappearance in a nucleus (Fig. 8). In the minimum spanning tree diagram, two directional trends are visible: nucleoli in demethylated main and lateral roots (mrd, lrd) are closer to “uninucleolar state” (a greater tendency to fusion of nuclei), and nucleoli in nuclei with methylated (control) genomes (mrc, lrc) are closer to “anucleolar state” (a greater tendency of nucleoli to be not fused)—see Fig. 8.
A minimum spanning tree (MST) of OTUs (operational taxonomic units) described by characteristics of nucleolar behaviour for main (mr) and lateral roots (lr), demethylated (d) or from control plants (c) with native genomes. For nucleolar behaviour characteristics, a matrix of generalised taxonomic distances was calculated and used in the non-metric multidimensional scaling method according to Rohlf (1994). Trends of the nucleolar fusion are shown by arrows. The tree is located in the space of three ordination axes (x,y,z)
Nine patterns have been documented for the fusion of the two main NORs located on 1B and 6B chromosomes in common wheat cv. Chinese Spring. Nucleolar fusion appeared to be non-random and dependent on NOR position in a nucleus (Jordan et al. 1982). The number of patterns would undoubtedly be increased if minor NORs in A and D wheat genomes were also taken into account. The number of nucleolar fusion patterns is correlated with the level of ploidy and its frequency is always presented in the form of leptokurtic and positively skewed curves (Kosina 1996).
For common wheat, Mukai et al. (1991) proved that rDNA sequences are mostly inactive in the form of facultative heterochromatin and only a part is decondensed and active. The rDNA decondensation pattern appears to be species-specific and varies between wheat and rye (Leitch et al. 1992). Changes in chromatin condensation have also been noted in a wheat translocation line with a 1R rye chromosome after 5-azaC application (Glyn et al. 1997). It is known that the nucleolar volume in wheat depends strictly on the size of the NOR constriction (Martini and Flavell 1985). The size and activity of nucleoli, detected as RNA by DAPI and propidium iodide staining, has been increased in a Triticum timopheevii Zhuk. × Aegilops umbellulata Zhuk. amphiploid after treatment of germinating seeds with 5-azaC (Kosina and Markowska 2010).
The potential for rDNA genes expression (Ag-NOR) varies between plant organs and tissues. In Brassica L. species, Chen and Pikaard (1997) found that expression of rRNA genes occurs in floral organs, while they are silenced in vegetative tissues. No maternal or ploidy factors change the expression of these genes. The developmental dependence of methylation in Brassica is also corroborated by methylation patterns in Triticale (Amado et al. 1997). Hasterok and Maluszynska (2000) proved that the number of NOR-loci is different in primary and adventitious roots in Allium cepa L. In the adventitious roots, two loci appeared to be minor and suppressed. Their rRNA potential was lower than that in primary roots. Kosina and Markowska (2011) reported that various patterns of nucleoli fusion in main and lateral roots in a Triticum timopheevii × Aegilops umbellulata amphiploid had been changed after 5-azaC treatment, and demethylation increased the number of these patterns, up to sixty – more NORs leads to more association patterns. In aneuploidal forms and amphiploids, sometimes expressing cytogenetic instability, the NOR variation is enhanced.
In addition, patterns of rDNA methylation in different accessions of Hordeum spontaneum (K. Koch) Thell. (wild barley) can be environment dependent and varies in microniches occupied by different accessions of the species (Sharma et al. 2005).
The number of rDNA loci is always higher than the numbers of NORs, and no nucleolar dominance has been noted in allotetraploid Brassica derivatives versus their progenitors (Hasterok and Maluszynska 2000). A different, parent-dependent pattern of rDNA expression has been discovered in Brachypodium L. 45S rDNA loci from one parent are suppressed in allotetraploid Brachypodium, while loci of diploid B. distachyon are always active (Idziak and Hasterok 2008). Carvalho et al. (2010) discovered that accessions of common wheat presenting the expression of four instead of six NORs have some loci which are more methylated. Variation in the number of NORs also depends on the stability of minor rDNA loci. They can be lost or expressed instead of deleted major ones. For instance, loss or suppression of NOR loci has been noted in barley (Nicoloff et al. 1977), Festuca arundinacea Schreb. (Thomas et al. 1997) and in Zingeria trichopoda (Boiss.) P.A. Smirn. (Kotseruba et al. 2003). Such a cytogenetic change can occur during structural rearrangements of chromosomes or can be induced during polyploidisation. Numerous minor NORs detected in hexaploid oats (Badaeva et al. 2011) will create broad nucleolar variation in their hybrid progeny, which is also clearly presented in Fig. 7.
Structural variation
Germination and growth
Significant differences between demethylated amphiploid plants and those untreated have been observed in changes of germination patterns (Fig. 9A, B) and plant growth dynamics (Fig. 10). The 5-azaC treated caryopses expressed slower germination. Growth of coleoptiles and main embryonic roots was slower and after 4 days of germination the lateral roots were not developed (Fig. 9B). The height of demethylated plants was lower than that of untreated plants during the 3 weeks of seedling growth (Fig. 10), and this difference between control and 5-azaC treated plants was maintained until the maturity of the plant.
The above results relate well to demethylation experiments with Linum usitatissimum L. Demethylation induces the development of poor seeds, probably due to shortening of the flowering time, and decreases the height of flax plants (Fieldes 1993; Fieldes and Amyot 1999). In contrast, in Triticale demethylation increases the height of plants, the number of tillers and delays maturity, irrespectively of 5-azaC concentration from 0.1 to 2.0 mM (Heslop-Harrison 1990). This means that the vegetative phase was strengthened, but not the generative one (compare poor germination and slower growth in the Avena amphiploid and flax).
In the Avena amphiploid, demethylation reduces pollen grains viability, especially in the second flower of the spikelet, and increases the number of multiporate pollen grains. The inter-plant variation of pollen grains characteristics is larger for demethylated plants than for those with native genomes (Florek 2013).
A demethylation experiment conducted in another amphiploid, Triticum timopheevii × Aegilops umbellulata, proved that the gross morphology of spike and spikelet exhibited a broader intra- and inter-plant variation under this treatment (Kosina and Markowska 2010).
Liu et al. (1999) study on hybrid introgression lines in rice proved that patterns of DNA methylation are very flexible and these were observed as hypermethylation or hypomethylation, which changed many plant characteristics.
Thus, methylation appeared as a very dynamic process, and changes induced by demethylation are more of a quantitative than a qualitative nature. The influence of demethylation on plant growth, flowering time and seed set in flax (Fieldes 1993) and analysis of Arabidopsis endosperm mutants (Grossniklaus et al. 2001) corroborates this point of view.
Variation of caryopsis structure
In grass caryopses, especially in cereals, above the nucellar projection towards the dorsal part of the seed, a special structure called the “endosperm cavity” is formed. In oats, an endosperm cavity can be T- or I-shaped (Florek 2013). The T-shaped cavity is more frequent in Avena barbata and the I-shaped is typical for A. sativa subsp. nuda. The I-shaped cavity dominates in the amphiploid. The I-cavity in untreated plants occurs with a frequency of 80%. Such a cavity can also be developed in the form of a long structure reaching the dorsal aleurone layer, and it occurs in 24% of seeds (Fig. 11A). The I-cavity is noted for 90% of the 5-azaC caryopses. The frequency of a deep I-cavity elongated between the ventral and dorsal aleurone layers is distinctly increased (to 40%) in treated 5-azaC caryopses. The long cavity divides a starchy endosperm into two (Fig. 11A) or three macrodomains (Fig. 11B). The development of the 5-aza long I-cavity is correlated with a poorly developed starchy endosperm in the dorsal part of the caryopsis. The number of cytokineses is decreased there.
Anomalies in the development of endosperm induced by 5-azaC treatment. A, B Overgrowth of I-cavities (white lines) dividing the endosperm tissue into two or more domains; C a clone of highly polyploidised cells (black dots) in an internal starchy endosperm; D collapsed starchy cells defective in starch synthesis (white arrow) and synthesising hemicelluloses in parts of thick cell walls (black arrow); E callus-like multiplied aleurone cells in the aleurone layer (encircled by the dotted line); F an anomalous location of starch synthesis in the dorsal area of a caryopsis, between the aleurone layer and testa (black arrow)
The development of large, polyploid starchy cells is a typical change caused by 5-azaC treatment (Fig. 11C). Other rare anomalies noted in caryopses of the demethylation origin are:
-
1.
Poor cell filling by starch granules in the central endosperm. Such a tissue is collapsed, while adjacent cells develop thick hemicellulosic walls (Fig. 11D).
-
2.
The local multiplication of aleurone cells (Fig. 11E).
-
3.
The synthesis of starch granules in the dorsal part of the caryopsis, in the outer space situated between the aleurone layer and testa (Fig. 11F).
Single cells (Fig. 12C, D), or groups of large polyploid (Fig. 12E) or smaller cells (Fig. 12B) were observed in isolated aleurone layers in both types of caryopses, with native genomes or demethylated. Thus, in the aleurone layer, the cell cycle can be elongated in large cells or shortened in small ones. Sometimes groups of small aleurone cells developed during a short cell cycle are lighter, because they do not contain aleurone grains (”a-aleurone” cells in Fig. 12B). The polyploid aleurone cells developed in the aleurone layer can synthesise thick hemicellulosic walls (Fig. 12D). A starchy cell phenotype within the aleurone layer is rarely developed (Fig. 12E). A new cell phenotype was noted in the aleurone layer of the demethylated caryopses (Fig. 12F). This phenotype is expressed in the form of long “a-aleurone” cells, growing in a tangential direction like fibres or sclereids. Such cells are empty and assimilates are stored in their thick walls (Kosina et al. 2014).
Anomalous “demethylation endosperm phenotypes” in the Avena amphiploid. A An extensive area of defective endosperm seen on the surface of the isolated aleurone layer. Groups of cells of various ploidy levels and parts empty of cells are visible. B A clone of cells free of aleurone grains (lighter cells) and presenting shortened cell cycle versus adjacent aleurone cells; C a highly polyploidised aleurone cell presenting an extended cell cycle, and a similar one in D with thick hemicellulosic walls (black arrow); E a starchy cell phenotype within an aleurone layer (black arrow) and a large clone of highly polyploidised aleurone cells (white arrow); F a rare “fibre-sclereid-like” aleurone phenotype of two cells (white arrows) which are empty of aleurone grains and express an intrusive growth in tangential direction
In addition, a distinct difference is observed in the frequency of developmental deviations between both states: caryopses with native versus demethylated genomes. The aleurone layer with large anomalous fragments (Fig. 12A) occurs with a 24% frequency in caryopses with native genomes, while demethylation increases their frequency to 59%. All the above developmental events form a kind of tissue mosaic, either inside the starchy endosperm or outside within the aleurone layer.
Ivanovskaya (1983) proved that the first endosperm cells in Triticum L./Elymus L. amphiploids are initiated in distinct areas at the wall of the embryo sac, which are separated from each other. Later, the cytokinetic activity forms endospermal clones. This developmental pattern created by cellular and free-cellular areas opens the possibility for the diversified development of the former. Particularly at the borders of these cellular groups, the intrusive growth of aleurone or nucellar tissues separates large endospermal domains (Kosina et al. 2013a), also shown in Figs. 10A, B and 11A. Such domains are also characteristic in an amphiploid Triticum timopheevii × Aegilops umbellulata, in both genomic states: native and demethylated (Kosina et al. 2013b). Deviations in the caryopsis development have been frequently noted in grass hybrids (Sabelli and Larkins 2009). Also, Fieldes and Amyot (1999) found that application of 5-azaC induced development of few poor seeds in the flax capsule. In plant hybrids, the expression of endosperm genes of two-parent origin is regulated by epigenetic imprinting and this phenomenon stabilises the tissue development (Sabelli and Larkins 2009).
Cytokineses in the embryo sac are active at its wall during the entire development of a caryopsis. In endosperm, older cells are pushed to the centre of the embryo sac by sequential periclinal divisions. Finally, a conical structure is formed, with the youngest cells in the aleurone layer. Normally, the central large part of the embryo sac is filled with starchy cells; however, sometimes the synthesis of starch is far ahead of cytokineses and the interior of the embryo sac is formed as a starchy “a-cellular” structure. Such a development has been noted in various grasses: Bromus secalinus L. (Skowrońska 2005); Avena strigosa Schreb. and A. brevis Roth. (Grabińska 2008), and Aegilops umbellulata (Koźlik 2013). In Fig. 10D demethylation caused the reverse situation, where starch synthesis was defective and delayed versus the cytokinetic activity. The central cells are poorly filled with starch and are partly collapsed.
The older central cells enter into endoreduplication cycles and form highly polyploidal units. In interploidy crosses, in Zea mays L., after six endoreduplication cycles, nuclei were changed by apoptosis; however, endoreduplication is delayed at a surplus of paternal genomes (Leblanc et al. 2002). Undoubtedly, apoptosis plays a selective role in endosperm via the elimination of abnormal tissue units, and creates new paths for growth of normal cells. Figure 10C proves that highly polyploidal cells can be developed near the aleurone layer, and a similar development has been observed in endosperm of the species Brachypodium distachyon (Kosina et al. 2012b) and an amphiploid Triticum timopheevii × Aegilops umbellulata (Koźlik 2013). Adjacent endosperm cells are compressed and changed apoptotically in favour of a polyploidal unit. This means that polyploidal, endoreduplicated cells can be “aggressive” against surroundings and cannot be apoptotised.
Alterations in endosperm development can relate to various aspects such as: metabolism (starch—Fig. 10D, hemicelluloses—Fig. 11D aleurone grains—Fig. 11B); shortening or lengthening of the cell cycle—Fig. 11B-D; amplification of aleurone cytokineses—Fig. 10E; change of the morphogenetic path of aleurone cells into periclinal “fibres”—Fig. 11F. The synthesis of starch granules in the space between the dorsal aleurone layer and testa is unique and is observed after demethylation. Starch synthesis outside the embryo sac, in the ventral endosperm cavity, has been observed in Avena strigosa (Grabińska 2008) and common wheat (Kosina 1979). In wheat this trait is cultivar-specific (Kosina et al. 2012a). Starch storage in the endosperm cavity can be associated with some discontinuity of the aleurone layer in the crease of the caryopsis. Such discontinuity can also be easily formed in the dorsal part of the caryopsis, where many aleurone domains are loosely connected (see Fig. 11A), or can be a remnant of the metabolic activity of the nucellus. The above phenomena prove that starch synthesis exceeds the capability of endosperm cellularization.
The amplification of cells in the aleurone layer (see Fig. 10E) was also noted by Lid et al. (2004) in disorgal 1 and disorgal 2 mutants in maize. These mutations are also related to changes noted by authors in root and leaf epidermis. Such amplification of the aleurone layer has been observed in Brachypodium retusum (Pers.) P. Beauv. (Kłyk 2005) and in an amphiploid Triticum timopheevii × Aegilops umbellulata with native or demethylated genomes (Koźlik 2013).
Concluding remarks
The 5-azaC demethylating experiments conducted with a cytogenetically more stable octoploid selection obtained from the decaploid Avena amphiploid proved that in its genomes are sites more resistant to demethylating agents. Increased concentrations of 5-azaC caused increased demethylation of genomes; however, subtelomeric/telomeric regions, some intercalary segments (C-bands?) and some chromosomes remained methylated. These chromosomes probably belong to the highly heterochromatic C genome. In the amphiploid, NORs and 5S rDNA loci were situated on five pairs of chromosomes, on three chromosomes both kinds of loci were colocalised. The chromosomes were of A/D, C and less frequently of B genome origin. The B genome seems to be less stable in the amphiploid. After 5-azaC demethylation one pair of minor NORs was activated. Minor loci of 25S rDNA were more effectively demethylated, while major loci can probably increase their own activation. Demethylation caused different nucleolar behaviours in main versus lateral roots. The trend towards fusion of nucleoli was marked under 5-azaC treatment, while in roots with native, methylated genomes nuclei were more multi-nucleolar. Demethylation reduced the growth of plants and the ability of seeds to germinate. The most remarkable changes induced by demethylation of the amphiploid genomes were noted in the endosperm development, because this tissue was in the native state more methylated, as has been proved for other species. In the amphiploid caryopses, the endosperm cavity was developed in the form of a paternal one. The cavity often divided endosperm tissue into several domains in caryopses with demethylated genomes. Also, multiple macrodisorders in the development of starchy and aleurone endosperm occurred. In addition, defects in starch synthesis and an increased level of the ploidy of starchy and aleurone cells were induced by 5-azaC. New cell and tissue “demethylation” phenotypes in endosperm were discovered. All the presented data prove that demethylation affected development in a quantitative rather than qualitative manner.
References
Amado L, Abranches R, Neves N, Viegas W (1997) Development-dependent inheritance of 5-azacytidine-induced epimutations in triticale: analysis of rDNA expression patterns. Chromosome Res 5:445–450
Antequera F, Bird A (1993) DNA methylation and CpG islands. In: Heslop-Harrison JS, Flavell RB (eds) The chromosome. BIOS Scientific Publishers Ltd., Oxford, pp 127–133
Armstrong KC, Le H, Fedak G (1991) Expression of Thinopyrum distichum NORs in wheat × Thinopyrum amphiploids and their backcross generations. Theor Appl Genet 81:363–368
Badaeva ED, Shelukhina OY, Dedkova OS (2011) Comparative cytogenetic analysis of hexaploid Avena L. species. Russ J Genet 47:691–702
Borowska N, Idziak D, Hasterok R (2011) DNA methylation patterns of Brachypodium distachyon chromosomes and their alteration by 5-azacytidine treatment. Chromosome Res 19:955–967
Carvalho A, Polanco C, Lima-Brito J, Guedes-Pinto H (2010) Differential rRNA genes expression in hexaploid wheat related to NOR methylation. Plant Mol Biol Rep 28:403–412
Castilho A, Neves N, Rufini-Castiglione M, Viegas W, Heslop-Harrison JS (1999) 5-methylcytosine distribution and genome organization in triticale before and after treatment with 5-azacytidine. J Cell Sci 112:4397–4404
Chen Q, Armstrong K (1994) Genomic in situ hybridization in Avena sativa. Genome 37:607–612
Chen ZJ, Pikaard CS (1997) Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. P Nat Acad Sci USA 94:3442–3447
Cremonini R, Ruffini Castiglione M, Grif VG, Kotseruba VV, Punina EO, Rodionov AV, Muravenko OV, Popov KV, Samatadze TE, Zelenin AV (2003) Chromosome banding and DNA methylation patterns, chromatin organisation and nuclear DNA content in Zingeria biebersteiniana. Biol Plant 46:543–550
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Fieldes MA (1993) Heritable effects of 5-azacytidine treatments on the growth and development of flax (Linum usitatissimum) genotrophs and genotypes. Genome 37:1–11
Fieldes MA, Amyot LM (1999) Evaluating the potential of using 5-azacytidine as an epimutagen. Can J Bot 77:1617–1622
Florek MZ (2013) Zmienność strukturalna i cytogenetyczna amphidiploida Avena barbata x Avena nuda o genomach natywnych i demetylowanych. Ph.D. thesis, University of Wrocław
Friebe B, Endo TR, Gill BS (1996) Chromosome-banding methods. In: Fukui K, Nakayama S (eds) Plant chromosomes: laboratory methods. CRC Press, Boca Raton, pp 123–153
Gehring M, Henikoff S (2007) DNA methylation dynamics in plant genomes. Biochim Biophys Acta 1769:276–286
Gerlach WL, Dyer TA (1980) Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res 11:4851–4865
Glyn MCP, Egertová M, Gazdova B, Kovarik A, Bezdek M, Leitch AR (1997) The influence of 5-azacytidine on the condensation of the short arm of rye chromosome 1R in Triticum aestivum L. root tip meristematic nuclei. Chromosoma 106:485–492
Grabińska A (2008) Zmienność mikrostrukturalna owocu diploidalnych gatunków rodzaju Avena L. MSc thesis. University of Wrocław
Grant V (1981) Plant speciation. Columbia University Press, New York
Grossniklaus U, Spillane C, Page DR, Köhler C (2001) Genomic imprinting and seed development: endosperm formation with and without sex. Curr Opin Plant Biol 4:21–27
Hasterok R, Maluszynska J (2000) Different rRNA gene expression in primary and adventitious roots of Allium cepa L. Folia Histochem Cytobiol 38:181–184
Heslop-Harrison JS (1990) Gene expression and parental dominance in hybrid plants. Development 108: 21–28
Heslop-Harrison JS (2000) Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell 12:617–635
Holden JHW (1966) Species relationships in the Aveneae. Chromosoma 20:75–124
Hsieh T-F, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324:1451–1454
Idziak D, Hasterok R (2008) Cytogenetic evidence of nucleolar dominance in allotetraploid species of Brachypodium. Genome 51:387–391
Irigoyen ML, Loarce Y, Linares C, Ferrer E, Leggett M, Fominaya A (2001) Discrimination of the closely related A and B genomes in AABB tetraploid species of Avena. Theor Appl Genet 103:1160–1166
Ivanovskaya EV (1983) Citoembriologicheskoe issledovanie differencirovki kletok rastenij. Izdatel’stvo Moskovskogo Universiteta, Moskva
Jellen EN, Rooney WL, Phillips RL, Rines HW (1993) Characterization of the hexaploid oat Avena byzantina cv. Kanota monosomic series using C-banding and RFLPs. Genome 36:962–970
Jellen EN, Gill BS, Cox TS (1994a) Genomic in situ hybridization differentiates between A/D- and C-genome chromatin and detects intergenomic translocations in polyploid oat species (genus Avena). Genome 37:613–618
Jellen EN, Phillips RL, Rines HW (1994b) Chromosomal localization and polymorphisms of ribosomal DNA in oat (Avena spp.). Genome 37:23–32
Jenkins G, Hasterok R (2007) BAC ‘landing’ on chromosomes of Brachypodium distachyon for comparative genome alignment. Nat Protocols 2:88–98
Jordan EG, Martini G, Bennett MD, Flavell RB (1982) Nucleolar fusion in wheat. J Cell Sci 56:485–495
Katsiotis A, Hagidimitriou M, Heslop-Harrison JS (1997) The close relationship between the A and B genomes in Avena L. (Poaceae) determined by molecular cytogenetic analysis of total genomic, tandemly and dispersed repetitive DNA sequences. Ann Bot 79:103–109
Kłyk B (2005) Zmienność mikrostrukturalna niektórych gatunków rodzaju Brachypodium P. Beauv. PhD thesis, University of Wrocław
Kosina R (1979) Association between structure and quality of the wheat grain. Cereal Res Commun 7:11–17
Kosina R (1996) Nucleolar variation in grass endosperm. Acta Soc Bot Pol 65:190
Kosina R, Heslop-Harrison JS (1996) Molecular cytogenetics of an amphiploid trigeneric hybrid beetwen Triticum durum, Thinopyrum distichum and Lophopyrum elongatum. Ann Bot 78:583–589
Kosina R, Markowska K (2010) Patterns of variation in Triticum timopheevii × Aegilops umbellulata amphiploid after demethylation of genomes. Annu Wheat Newsl 56:207–208
Kosina R, Markowska K (2011) Nucleolar variability in Triticum timopheevii × Aegilops umbellulata amphiploid. Annu Wheat Newsl 57:253
Kosina R, Tomaszewska P, Kamińska K (2012a) On caryopsis crease and endosperm cavity in wheat and Brachypodium distachyon. Annu Wheat Newsl 58:196–197
Kosina R, Tomaszewska P, Kamińska K (2012b) On caryopsis developmental events in wheat and Brachypodium distachyon. Annu Wheat Newsl 58:197–198
Kosina R, Bureś MK, Florek M, Grabińska A, Kawa P, Kłyk B, Kochmański Ł, Koźlik A, Kurek A, Skowrońska J, Tomaszewska P, Zając D (2013a) Structural characteristics of grass hybrid endosperm development. Annu Wheat Newsl 59:116–117
Kosina R, Koźlik A, Markowska K (2013b) Endospermal domains in a ‘Triticum timopheevii subsp timopheevii/Aegilops umbellulata’ amphiploid. Annu Wheat Newsl 59:117–118
Kosina R, Tomaszewska P, Florek M (2014) Microstructural variability of endosperm in Triticeae versus Avena. Annu Wheat Newsl 60:118–119
Kotseruba V, Gernand D, Meister A, Houben A (2003) Uniparental loss of ribosomal DNA in the allotetraploid grass Zingeria trichopoda (2n = 8). Genome 46:156–163
Koźlik A (2013) Zmienność mikrostrukturalna owocu amfiploida Triticum timopheevii × Aegilops umbellulata. MSc thesis, University of Wrocław
Kushwaha N, Zadoo SN, Choubey RN (2004) Breakdown of polyploidy and isolation of stable polysomics in amphidecaploid Avena sativa L. × A. maroccana Gdgr. Cytologia 69:41–47
Lacadena JR, Cermeño MC, Orellana J, Santos JL (1988) Nucleolar competition in Triticeae. In: Brandham PE (ed) Kew chromosome conference III. HMSO, London, pp 151–165
Leblanc O, Pointe C, Hernandez M (2002) Cell cycle progression during endosperm development in Zea mays depends on parental dosage effects. Plant J 32:1057–1066
Leitch AR, Mosgöller W, Shi M, Heslop-Harrison JS (1992) Different patterns of rDNA organization at interphase in nuclei of wheat and rye. J Cell Sci 101:751–757
Levy AA, Feldman M (2002) The impact of polyploidy on grass genome evolution. Plant Physiol 130:1587–1593
Lewis J, Bird A (1991) DNA methylation and chromatin structure. FEBS Lett 285:155–159
Lid SE, Al RH, Krekling T, Meeley RB, Ranch J, Opsahl-Ferstad H-G, Olsen O-A (2004) The maize disorganized aleurone layer 1 and 2 (dil1, dil2) mutants lack control of the mitotic division plane in the aleurone layer of developing endosperm. Planta 218:370–378
Linares C, González J, Ferrer E, Fominaya A (1996) The use of double fluorescence in situ hybridization to physically map the positions of 5S rDNA genes in relation to the chromosomal location of 18S–5.8S–26S rDNA and a C genome specific DNA sequence in the genus Avena. Genome 39:535–542
Liu B, Wendel JF (2003) Epigenetic phenomena and the evolution of plant allopolyploids. Mol Phylogenet Evol 29:365–379
Liu B, Piao H, Zhao F, Zhao J, Liu Z, Huang B (1999) DNA methylation changes in rice induced by Zizania latifolia (Griseb.) DNA introgression. Hereditas 131:75–78
Majerová E, Fojtová M, Mozgová I, Bittová M, Fajkus J (2011) Hypomethylating drugs efficiently decrease cytosine methylation in telomeric DNA and activate telomerase without affecting telomere lengths in tobacco cells. Plant Mol Biol 77:371–380
Martini G, Flavell RB (1985) The control of nucleolus volume in wheat, a genetic study at three developmental stages. Heredity 54:111–120
Messeguer R, Ganal MW, Steffens JC, Tanksley SD (1991) Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol Biol 16:753–770
Mukai Y, Endo TR, Gill BS (1991) Physical mapping of the 18S. 26S rRNA multigene family in common wheat: identification of a new locus. Chromosoma 100:71–78
Nicoloff H, Anastassova-Kristeva M, Künzel G, Rieger R (1977) The behavior of nucleolus organizers in structurally changed karyotypes of barley. Chromosoma 62:103–109
Nikoloudakis N, Katsiotis A (2008) The origin of the C-genome and cytoplasm of Avena polyploids. Theor Appl Genet 117:273–281
Nishiyama I (1962) Cytogenetic studies in Avena, IX. New synthetic oats in the progenies of induced decaploid interspecific hybrids. Jap J Genet 37:118–130
Nishiyama I, Tabata M, Iizuka M (1963) Cytogenetic studies in Avena, XI. Chromosomal constitution of the inbred progeny of synthetic amphiploid oats (10×). Jap J Genet 38:123–134
Phillips RL, Rines HW, Jellen EN, Rooney WL, Wu BX, Kianian S, Riera-Lizarazu O (1995) Oat genome analysis via molecular markers and oat × corn crosses. In: Raupp WJ, Gill BS (eds) Classical and molecular cytogenetic analysis. Kansas Agricultural Research Station Report 95-352-D, pp 49–57
Pikaard CS (2000) The epigenetics of nucleolar dominance. Trends Genet 16:495–500
Rohlf FJ (1994) NTSYS-pc, version 1.80. Exeter Software, New York
Ruffini Castiglione M, Cremonini R, Frediani M (2002) DNA methylation patterns on plant chromosomes. Caryologia 55:275–282
Sabelli PA, Larkins BA (2009) The development of endosperm in grasses. Plant Physiol 149:14–26
Schwarzacher T, Heslop-Harrison P (2000) Practical in situ hybridization. BIOS Scientific Publishers Limited, Oxford
Sharma S, Balyan HS, Gupta PK (2005) Adaptive methylation pattern of ribosomal DNA in wild barley from Israel. Barley Genet Newsl 35:27–35
Siroky J, Ruffini Castiglione M, Vyskot B (1998) DNA methylation patterns of Melandrium album chromosomes. Chromosome Res 6:441–446
Skowrońska J (2005) Mikrostrukturalna i cytogenetyczna analiza grupy Bromus secalinus–B. commutatus–B. racemosus. MSc thesis, University of Wrocław
Thomas HM, Harper JA, Meredith MR, Morgan WG, King IP (1997) Physical mapping of ribosomal DNA sites in Festuca arundinacea and related species by in situ hybridization. Genome 40:406–410
Tomaszewska P, Kosina R (2013) On the different role of parental genomes in selected hybrid grasses. Chromosome Res 21(Suppl 1):131–132
Tomaszewska P, Kosina R (2015) The A, B, C and D genomes in endosperm of oat amphiploids. Chromosome Res 23(Suppl. 1):128
Unfried I, Gruendler P (1990) Nucleotide sequence of the 5.8 S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res 18:4011
Yang Q, Hanson L, Bennett MD, Leitch IJ (1999) Genome structure and evolution in the allohexaploid weed Avena fatua L. (Poaceae). Genome 42:512–518
Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D (2010) Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA 107:18729–18734
Acknowledgements
This research was supported by the National Committee for Scientific Research (KBN) in Poland, Grant 4427/PB/IBR/10. The authors are very grateful to Prof. Robert Hasterok for donating the 25S and 5S rDNA probes.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Florek, M., Kosina, R. rDNA cytogenetics and some structural variability in an Avena barbata Pott ex Link × A. sativa subsp. nuda (L.) Gillet et Magne amphiploid after 5-azaC treatment. Genet Resour Crop Evol 64, 1723–1741 (2017). https://doi.org/10.1007/s10722-016-0469-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-016-0469-x