Abstract
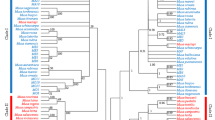
The ITS1-5.8S-ITS2 region of the 18S-28S nrDNA was sequenced in 49 Vicia species and subspecies selected from nine sections of the two subgenera to resolve taxonomic contradictions. Phylogenies derived from the internal transcribed spacer (ITS) sequences strongly support the view that both Vicia faba and Vicia bithynica are distant from the Narbonensis species complex (NSC) in section Faba. V. faba is more closely aligned with sections Peregrinae and Hypechusa, whereas V. bithynica is more closely aligned with section Vicia. Six synapomorphic substitutions unique to the NSC support a separate section for the Narbonensis complex. Vicia johannis subsp. johannis was the most diverged species within the NSC with high genetic distance between V. johannis subsp. johannis and remaining species and subspecies of the NSC. These results suggest that Vicia johannis subsp. johannis should be given species rank. The neighbor-joining and parsimony based trees supported the sections Vicia (after the exclusion of Vicia lathyroides) and Cracca (after the exclusion of Vicia vicioides and Vicia hirsuta). The section Hypechusa is polyphyletic and paraphyletic based on the present ITS sequence cladistic analysis. The x = 7 as the ancestral number for the genus is supported by the present study.






Similar content being viewed by others
References
Ainouche A-K, Bayer RJ (1999) Phylogenetic relationships in Lupinus (Fabaceae: Papilionoideae) based on internal transcribed spacers sequences (ITS) of nuclear ribosomal DNA. Am J Bot 86:590–607
Allan GJ, Porter JM (2000) Tribal delimitation and phylogenetic relationships of Loteae and Coronilleae (Faboideae: Fabaceae) with special reference to Lotus: evidence from nuclear ribosomal ITS sequence. Am J Bot 87:1871–1881
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the compositae. Mol Phylogenet Evol 1:3–16
Baldwin BG (1993) Molecular phylogenetics of Calycadenia (Compositae) based on ITS sequences of nuclear ribosomal DNA: chromosomal and morphological evolution reexamined. Am J Bot 80:222–238
Baldwin BG, Sanderson MJ, Porter JM, Woiciechowski ME, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Mo Bot Gard 82:247–277
Ball PW (1968) Vicia L.. In: Tutin TG et al (eds) Flora europaea, vol 2. Cambridge University Press, Cambridge, pp 129–136
Baum BR, Estabrook GF (1996) Impact of outgroup inclusion on estimates by parsimony of undirected branching of ingroup phylogenetic lines. Taxon 45:243–257
Bennett SJ, Maxted N (1997) An ecogeographic analysis of the Vicia narbonensis complex. Genet Resour Crop Evol 44:411–428
Caputo P, Frediani M, Venora G, Ravalli C, Ambrosio M, Cremonini R (2006) Nuclear DNA contents, rDNA, and karyotype evolution in subgenus Vicia: III. The heterogeneous section Hypechusa. Protoplasma 228:167–177
Choi BH, Kim JH (1997) ITS sequences and speciation on Far Eastern Indigofera (Leguminosae). J Plant Res 110:339–346
Chooi WY (1971) Variation in nuclear DNA content in the genus Vicia. Genetics 68:195–211
Davis PH, Plitman UP (1970) Vicia L. In: Davis PH (ed) Flora of Turkey and East Aegean Islands, vol 3. Edinburgh University Press, Edinburgh, pp 274–325
Endo Y, Choi BH, Ohashi H, Delgado-Salinas A (2008) Phylogenetic relationships of New World Vicia (Leguminosae) inferred from nrDNA internal transcribed spacer sequences and floral characters. Syst Bot 33(2):356–363
Fedoronchuk MM (1996) A revision of species of the genus Vicia (Fabaceae) in the flora of Ukraine. Ukr Bot J 53:587–597
Fennel SR, Powell W, Wright F, Ramsay G, Waugh R (1998) Phylogenetic relationships between Vicia faba (Fabaceae) and related species inferred from chloroplast trnL sequences. Plant Syst Evol 212:247–259
Frediani M, Caputo P, Venora G, Ravalli C, Ambrosio M, Cremonono R (2005) Nuclear DNA contents, rDNAs, and karyotype evolution in Vicia subgenus Vicia: II. Section Peregrinae. Protoplasma 226:181–190
Goel S, Raina SN, Ogihara Y (2002) Molecular evolution and phylogenetic implications of internal transcribed spacer sequences of nuclear ribosomal DNA in the Phaseolus-Vigna complex. Mol Phyl Evol 22:1–19
Hanelt P, Mettin D (1989) Biosystematics of the genus Vicia L. (Leguminosae). Annu Rev Ecol Syst 20:199–223
Hanelt P, Schäfer H, Schultze-Motel J (1972) Die Stellung von Vicia faba L. in der Gattung Vicia L. und Betrachtungen zu dieser Kulturart. Kulturpflanze 20:263–275
Harlan JR (1956) Theory and dynamics of grassland agriculture. Van Nostrand D Co. Inc., Princeton
Huang JC, Sun M (2000) Genetic diversity and relationships of sweetpotato and its wild relatives in Ipomea series Batatas (Convolvulaceae) as revealed by inter-simple sequence repeat (ISSR) and restriction analysis of chloroplast DNA. Theor Appl Genet 100:1050–1060
Jaaska V (1997) Isoenzyme diversity and phylogenetic affinities in Vicia subgenus Vicia (Fabaceae). Genet Resour Crop Evol 44:557–574
Jaaska V (2005) Isozyme variation and phylogenetic relationships in Vicia subgenus Cracca (Fabaceae). Ann Bot 96:1085–1096
Jaaska V (2008) Isozyme evidence on the specific distinctness and phylogenetic position of Vicia incisa (Fabaceae). Cent Eur J Biol 3(2):169–176
Kenicer GJ, Kajita T, Pennington RT, Murata J (2005) Systematics and biogeography of Lathyrus (Leguminosae) based on internal transcribed spacer and cpDNA sequence data. Am J Bot 92:1199–1209
Koul KK, Raina SN, Parida A, Bisht MS (1999) Sex differences in meiosis between Vicia faba L. and its close wild relatives. Bot J Linn Soc 129(3):239–247
Kupicha FK (1976) The infrageneric structure of Vicia. Notes R Bot Gard Edinb 34:287–326
Ladizinsky G (1975) Seed protein electrophoresis of the wild and cultivated species of selection Faba of Vicia. Euphytica 24:785–788
Ladizinsky G (1978) Chromosomal polymorphism in wild populations of Vicia sativa L. Caryologia 31:233–241
Leht M (2009) Phylogenetics of Vicia (Fabaceae) based on morphological data. Feddes Repert 120(7–8):379–393
Leht M, Jaaska V (2002) Cladistic and phonetic analysis of relationships in Vicia subgenus Vicia (Fabaceae) by morphology and isozymes. Plant Syst Evol 232:237–260
Martin Sanz A, Gilsanz Gonzalez S, Hasan Syed N, Jose Suso M, Caminero Saldana C, Flavell AJ (2007) Genetic diversity analysis in Vicia species using retrotransposon-based SSAP markers. Mol Genet Genomics 278:433–441
Maxted N (1992) Towards a faba bean progenitor!. FABIS Newslett 31:3–8
Maxted N (1993) A phenetic investigation of Vicia L. subgenus Vicia (Leguminosae, Vicieae). Bot J Linn Soc 111:155–182
Maxted N (1994) A phenetic investigation on Vicia section Peregrinae Kupicha (Leguminosae, Papilionoideae, Vicieae). Edinb J Bot 51:75–97
Maxted N (1995) An ecogeographical study of Vicia subgenus Vicia. Systematic and ecogeographic studies on crop genepools 8. International Plant Genetic Resources Institute, Rome, Italy. Available at: http://pdf.usaid.gov/pdf_docs/PNABU773.pdf
Maxted N, Douglas C (1997) A phonetic investigation of Vicia section Hypehusa (Alef.) Aschers. and Graebner (Leguminosae, Papilionoideae, Vicieae). Lagascalia 19:345–370
Maxted N, Callimassia MA, Bennett MD (1991) Cytotaxonomic studies of eastern Mediterranean Vicia species (Leguminosae). Plant Syst 177:221–234
Mayer MS, Bagga SK (2002) The phylogeny of Lens (Leguminosae): new insight from ITS sequence analysis. Plant Syst Evol 232:145–154
Mettin D, Hanelt P (1964) Cytosystematische Untersuchungen in der Artengruppe um Vicia sativa L. I. Kulturpflanze 12:163–225
Mettin D, Hanelt P (1973) Über Speziationsvorgänge in der Gattung Vicia L. Kulturpflanze 21:25–54
Mort ME, Soltis DE, Soltis PA, Ortega JF, Guerra AS (2002) Phylogenetics and evolution of the Macaronesian clade of Crassulaceae inferred from nuclear and chloroplast sequence data. Syst Bot 27(2):271–288
Padgett DJ, Les DH, Crow GEC (1999) Phylogenetic relationships in Nuphar (Nymphaeceae): evidence from morphology, chloroplast DNA, and nuclear ribosomal DNA. Am J Bot 86:1316–1324
Perrino P, Pignone D (1981) Contribution to the taxonomy of Vicia species belonging to the section Faba. Kulturpflanze 29:311–319
Plitmann U (1967) Biosystematical study of Vicia of the middle east. Private Publications, pp 1–128
Potokina E, Tomooka N, Vaughan DA, Alexandrova T, Xu RQ (1999) Phylogeny of Vicia subgenus Vicia (Fabaceae) based on analysis of RAPDs and RFLP of PCR-amplified chloroplast genes. Genet Resour Crop Evol 46:149–161
Potter D, Luby JJ, Harrison RE (2000) Phylogenetic relationships among species of Fragaria (Rosaceae) inferred from non-coding nuclear and chloroplast DNA sequences. Syst Bot 25:337–348
Przybylska J, Zimniak-Przybylska Z (1995) Electrophoretic seed albumin patterns and species relationships in Vicia sect. Faba (Fabaceae). Plant Syst Evol 198:179–194
Radzhi AD (1970) Conspectus systematis specierum caucasicarum generis Vicia L. In: Tzvelev NN (ed) Novitates systematicae plantarum vascularium, vol 6. Nauka, Leningrad, pp 228–240 (in Russian)
Raina SN (1990) Genome organization and evolution in the genus Vicia. In: Kawano S (ed) Biological approaches and evolutionary trends in plants. Acad Press, New York, pp 183–201
Raina SN, Bisht MS (1988) DNA amount and chromatin compactness in Vicia. Genetica 77:67–77
Raina SN, Narayan RKJ (1984) Changes in DNA composition in the evolution of Vicia species. Theor Appl Genet 68:187–192
Raina SN, Ogihara Y (1994) Chloroplast DNA diversity in Vicia faba and its close wild relative: implications for reassessment. Theor Appl Genet 88:261–266
Raina SN, Ogihara Y (1995) Ribosomal DNA repeat unit polymorphism in 49 Vicia species. Theor Appl Genet 90:47–486
Raina SN, Rees H (1983) DNA variation between and within chromosome complement of Vicia species. Heredity 51:335–346
Raina SN, Yamamoto K, Murakami M (1989) Intraspecific hybridization and its bearing on chromosome evolution in Vicia narbonensis (Fabaceae). Plant Syst Evol 167:201–217
Raina SN, Mukai Y, Kawaguchi K, Goel S (2001) Physical mapping of 18S-5.8S-26S and 5S ribosomal RNA gene families in three important vetches (Vicia species) and their allied taxa constituting three species complexes. Theor Appl Genet 103:839–845
Raina SN, Khandavilli K, Shiran B, Mahmoudi A (2010) Fluorescence in situ hybridization with multiple repeated DNA probes applied to the analysis of chromosomal and genomic relationship levels between faba bean (Vicia faba) and its close wild relatives. Caryologia 63(3):215–222
Ravash R, Shiran B, Alavi A, Bayat F, Rajaee S, Zervakis GI (2010) Genetic variability and molecular phylogeny of Pleurotus eryngii species-complex isolates from Iran, and notes on the systematics of Asiatic populations. Mycol Prog 9(2):181–194
Rieseberg LH, Soltis DE (1991) Phylogenetic consequences of cytoplasmic gene flow in plants. Evol Trends Plants 5:65–84
Roy SW (2009) Phylogenomics: gene duplication, unrecognized paralogy and outgroup choice. PLOS One 4(2):e 4568
Ruffini Castiglione M, Frediani M, Gelati MT, Ravalli C, Venora G, Caputo P, Cremonini R (2007) Cytological and molecular characterization of Vicia esdraelonensis Warb. and Eig: a rare taxon. Protoplasma 231:151–159
Ruffini Castiglione M, Frediani M, Gelati MT, Ravalli C, Venora G, Caputo P, Cremonini R (2011) Cytology of Vicia species. X. Karyotype evolution and phylogenetic implication in Vicia species of the sections Atossa, Microcarinae, Wiggersia and Vicia. Protoplasma 248:707–716
Ruffini Castiglione M, Frediani M, Gelati MT, Venora G, Giorgetti L, Caputo P, Cremonini R (2012) Cytological and molecular characterization of Vicia barbazitae Ten. and Guss. Protoplasma 249:779–788
Sakowicz T, Cieslikowski T (2006) Phylogenetic analyses within three sections of the genus Vicia. Cell Mol Biol Lett 11:594–615
Schaefer H, Hechenleitner P, Santos-Guerra A, de Sequeira MM, Pennington RT, Kenicer G, Carine MA (2012) Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-Atlantic island lineages. BMC Evol Biol 12(1):250
Schäfer HI (1973) Zur Taxonomie der Vicia narbonensis Gruppe. Kulturpflanze 21:211–273
Schubert I, Rieger R (1985) A new mechanism for altering chromosome number during karyotype evolution. Theor Appl Genet 70:213–221
Schubert I, Rieger R, Michaelis A (1986) Structural and numerical manipulation of the Vicia faba karyotype: Results and perspectives. Biol Zbl 105:9–17
Sehgal D, Rajpal VR, Raina SN (2008) Chloroplast DNA diversity reveals the contribution of the two wild species in the origin and evolution of diploid safflower (Carthamus tinctorius L.). Genome 51:638–643
Sehgal D, Raina SN, Devarumath RM, Sasanuma T, Sasakuma T (2009) Nuclear DNA assay in solving issues related to ancestry of the domesticated diploid safflower (Carthamus tinctorius L.) and the polyploid (Carthamus) taxa, and phylogenetic and genomic relationships in the genus Carthamus L. (Asteraceae). Mol Phylogenet Evol 53:631–644
Shiran B, Raina SN (2001) Evidence of rapid evolution and incipient speciation in Vicia sativa species complex based on nuclear and organellar RFLPs and PCR analysis. Genet Resour Crop Evol 48:519–532
Sonnante GI, Galasso I, Pignone D (2003) ITS sequence analysis and phylogenetic inference in the genus Lens Mill. Ann Bot 91:49–54
Stankevich AK (1983) The genus Vicia L. and its position in the tribe Fabeae (Fabaceae). Bull Appl Bot Genet Plant Breed 79:3–10 (in Russian)
Strimmer K, von Haeseler A (1997) Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA 94:6815–6819
Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Massachusetts
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tzvelev NN (1987) Vicia L. In: Fedorov AA (ed) Flora of the European part of the USSR, 6. Nauka, Leningrad, pp 127–147
Van de Van M, Powell W, Ramsay G, Waugh R (1990) Restriction fragment length polymorphisms as genetic markers in Vicia L. Heredity 65:329–342
Van de Ven M, Powell W, Ramsay G, Waugh R, Phillips M, Duncan N (1993) Taxonomic relationships between Vicia faba and its relatives based on nuclear and mitochondrial RFLPs and PCR analysis. Theor Appl Genet 86:71–80
Van Droogenbroeck B, Kyndt T, Maertens I, Romeijn-Peeters E, Scheldeman X, Romero-Motochi JP, Van Damme P, Goetghebeur P, Gheysen G (2004) Phylogenetic analysis of the highland papayas (Vasconcellea) and allied genera (Caricaeae) using PCR-RFLP. Theor Appl Genet 108:1473–1486
Venora G, Blangiflorati S, Frediani M, Maggini F, Gelati MT, Ruffini Castiglione M, Cremonini R (2000) Nuclear DNA contents, rDNAs, chromatin organization, and karyotype evolution in Vicia sect. faba. Protoplasma 213:118–125
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Gelfand D, Sminsky J, White T (eds) PCR Protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Wojciechowski MF, Sanderson MJ, Baldwin BG, Donoghue MJ (1993) Monophyly of aneuploidy Astragalus (Fabaceae): evidence from nuclear ribosomal DNA internal transcribed spacer sequences. Am J Bot 80:711–722
Wojciechowski MF, Sanderson MJ, Steele KP, Liston A (2000) Molecular phylogeny of the “temperate herbaceous tribes” of Papilionoid legumes: a supertree approach. Adv legum syst 9:277–298
Yamamoto K (1986) Interspecific hybridization among Vicia narbonensis and its wild relatives. Biol Zbl 105:181–197
Zohary D, Hopf M (1973) Domestication of pulses in the Old World. Science 182:887–894
Acknowledgments
Grateful thanks are due to National Academy of Sciences, India and Shahrekord University, Iran for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shiran, B., Kiani, S., Sehgal, D. et al. Internal transcribed spacer sequences of nuclear ribosomal DNA resolving complex taxonomic history in the genus Vicia L.. Genet Resour Crop Evol 61, 909–925 (2014). https://doi.org/10.1007/s10722-014-0086-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-014-0086-5